|
Haplotypes
A haplotype (haploid genotype) is a group of alleles in an organism that are inherited together from a single parent. Many organisms contain genetic material (DNA) which is inherited from two parents. Normally these organisms have their DNA organized in two sets of pairwise similar chromosomes. The offspring gets one chromosome in each pair from each parent. A set of pairs of chromosomes is called diploid and a set of only one half of each pair is called haploid. The haploid genotype (haplotype) is a genotype that considers the singular chromosomes rather than the pairs of chromosomes. It can be all the chromosomes from one of the parents or a minor part of a chromosome, for example a sequence of 9000 base pairs or a small set of alleles. Specific contiguous parts of the chromosome are likely to be inherited together and not be split by chromosomal crossover, a phenomenon called genetic linkage. As a result, identifying these statistical associations and a few alleles of a specif ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplotype Estimation
In genetics, haplotype estimation (also known as "phasing") refers to the process of statistical estimation of haplotypes from genotype data. The most common situation arises when genotypes are collected at a set of polymorphic sites from a group of individuals. For example in human genetics, genome-wide association studies collect genotypes in thousands of individuals at between 200,000-5,000,000 SNPs using microarrays. Haplotype estimation methods are used in the analysis of these datasets and allow genotype imputation of alleles from reference databases such as the HapMap Project and the 1000 Genomes Project. Genotypes and haplotypes Genotypes measure the unordered combination of alleles at each locus, whereas haplotypes represent the genetic information on multiple loci that have been inherited together from an individual's parents. Theoretically the number of possible haplotypes equals to the product of allele numbers of each locus in consideration. Specially, most of the SN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Imputation (genetics)
In genetics, imputation is the statistical inference of unobserved genotypes. It is achieved by using known haplotypes in a population, for instance from the HapMap or the 1000 Genomes Project in humans, thereby allowing to test for association between a trait of interest (e.g. a disease) and experimentally untyped genetic variants, but whose genotypes have been statistically inferred ("imputed"). Genotype imputation is usually performed on SNPs, the most common kind of genetic variation. Genotype imputation hence helps tremendously in narrowing down the location of probably causal variants in genome-wide association studies, because it increases the SNP density (the genome size remains constant, but the number of genetic variants increases) and thus reduces the distance between two adjacent SNPs. Context In genetic epidemiology and quantitative genetics, researchers aim at identifying genomic locations where variation between individuals is associated with variation in traits of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplogroup
A haplotype is a group of alleles in an organism that are inherited together from a single parent, and a haplogroup (haploid from the , ''haploûs'', "onefold, simple" and ) is a group of similar haplotypes that share a common ancestor with a single-nucleotide polymorphism mutation. More specifically, a haplotype is a combination of alleles at different chromosomal regions that are closely linked and tend to be inherited together. As a haplogroup consists of similar haplotypes, it is usually possible to predict a haplogroup from haplotypes. Haplogroups pertain to a single line of descent. As such, membership of a haplogroup, by any individual, relies on a relatively small proportion of the genetic material possessed by that individual. Each haplogroup originates from, and remains part of, a preceding single haplogroup (or paragroup). As such, any related group of haplogroups may be precisely modelled as a nested hierarchy, in which each set (haplogroup) is also a subset of a si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
International HapMap Project
The International HapMap Project was an organization that aimed to develop a haplotype map (HapMap) of the human genome, to describe the common patterns of human genetic variation. HapMap is used to find genetic variants affecting health, disease and responses to drugs and environmental factors. The information produced by the project is made freely available for research. The International HapMap Project is a collaboration among researchers at academic centers, non-profit biomedical research groups and private companies in Canada, China (including Hong Kong), Japan, Nigeria, the United Kingdom, and the United States. It officially started with a meeting on October 27 to 29, 2002, and was expected to take about three years. It comprises three phases; the complete data obtained in Phase I were published on 27 October 2005. The analysis of the Phase II dataset was published in October 2007. The Phase III dataset was released in spring 2009 and the publication presenting the final re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
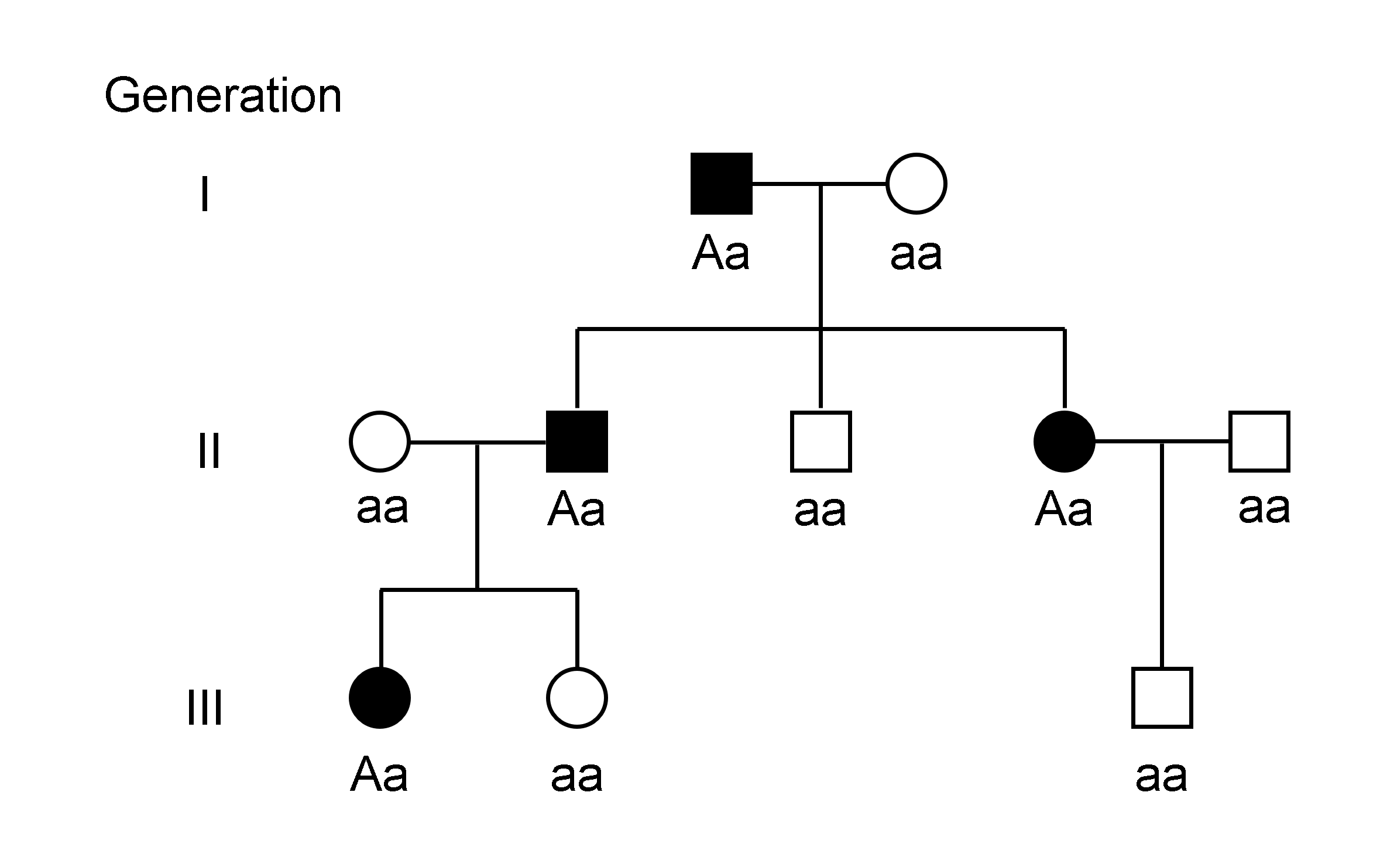
Genetic Linkage
Genetic linkage is the tendency of Nucleic acid sequence, DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two Genetic marker, genetic markers that are physically near to each other are unlikely to be separated onto different Chromatid, chromatids during chromosomal crossover, and are therefore said to be more ''linked'' than markers that are far apart. In other words, the nearer two Gene, genes are on a chromosome, the lower the chance of Genetic recombination, recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly ''unlinked'', although the penetrance of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located. Genetic linkage is the most prominent exception to Gregor M ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondrial DNA
Mitochondrial DNA (mtDNA and mDNA) is the DNA located in the mitochondrion, mitochondria organelles in a eukaryotic cell that converts chemical energy from food into adenosine triphosphate (ATP). Mitochondrial DNA is a small portion of the DNA contained in a eukaryotic cell; most of the DNA is in the cell nucleus, and, in plants and algae, the DNA also is found in plastids, such as chloroplasts. Mitochondrial DNA is responsible for coding of 13 essential subunits of the complex oxidative phosphorylation (OXPHOS) system which has a role in cellular energy conversion. Human mitochondrial DNA was the first significant part of the human genome to be sequenced. This sequencing revealed that human mtDNA has 16,569 base pairs and encodes 13 proteins. As in other vertebrates, the human mitochondrial genetic code differs slightly from nuclear DNA. Since animal mtDNA evolves faster than nuclear genetic markers, it represents a mainstay of phylogenetics and evolutionary biology. It als ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genotypes
The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a specific gene depends on the number of copies of each chromosome found in that species, also referred to as ploidy. In diploid species like humans, two full sets of chromosomes are present, meaning each individual has two alleles for any given gene. If both alleles are the same, the genotype is referred to as Zygosity, homozygous. If the alleles are different, the genotype is referred to as heterozygous. Genotype contributes to phenotype, the observable traits and characteristics in an individual or organism. The degree to which genotype affects phenotype depends on the trait. For example, the petal color in a pea plant is exclusively determined by genotype. The petals can be purple or white depending on the alleles present in the pea plan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
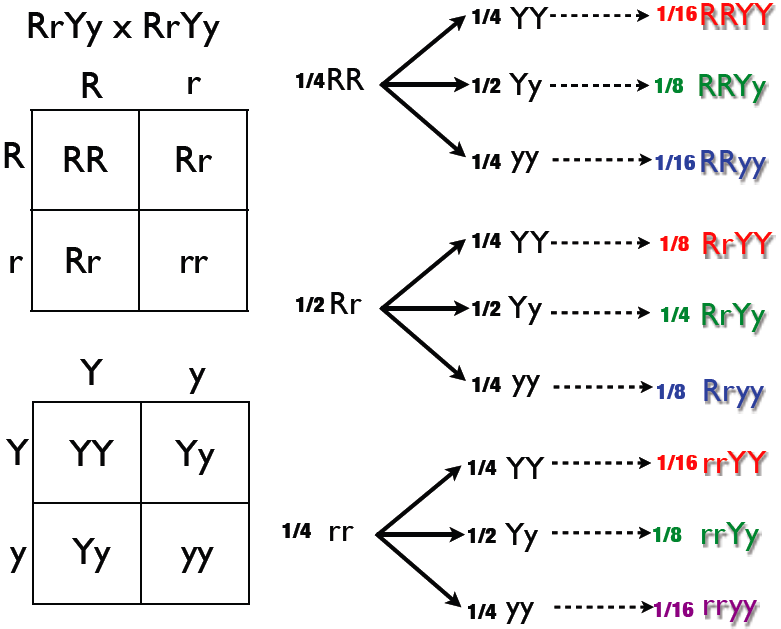
Punnett Square
The Punnett square is a square diagram that is used to predict the genotypes of a particular cross or breeding experiment. It is named after Reginald C. Punnett, who devised the approach in 1905. The diagram is used by biologists to determine the probability of an offspring having a particular genotype. The Punnett square is a tabular summary of possible combinations of maternal alleles with paternal alleles. These tables can be used to examine the genotypical outcome probabilities of the offspring of a single trait (allele), or when crossing multiple traits from the parents. The Punnett square is a visual representation of Mendelian inheritance, a fundamental concept in genetics discovered by Gregor Mendel. For multiple traits, using the "forked-line method" is typically much easier than the Punnett square. Phenotypes may be predicted with at least better-than-chance accuracy using a Punnett square, but the phenotype that may appear in the presence of a given genotype can in s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heterozygous
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism. Most eukaryotes have two matching sets of chromosomes; that is, they are diploid. Diploid organisms have the same locus (genetics), loci on each of their two sets of homologous chromosomes except that the sequences at these loci may differ between the two chromosomes in a matching pair and that a few chromosomes may be mismatched as part of a chromosomal Sex-determination system#Chromosomal determination, sex-determination system. If both alleles of a diploid organism are the same, the organism is #Homozygous, homozygous at that locus. If they are different, the organism is #Heterozygous, heterozygous at that locus. If one allele is missing, it is #Hemizygous, hemizygous, and, if both alleles are missing, it is #Nullizygous, nullizygous. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ambiguous
Ambiguity is the type of meaning in which a phrase, statement, or resolution is not explicitly defined, making for several interpretations; others describe it as a concept or statement that has no real reference. A common aspect of ambiguity is uncertainty. It is thus an attribute of any idea or statement whose intended meaning cannot be definitively resolved, according to a rule or process with a finite number of steps. (The prefix '' ambi-'' reflects the idea of " two", as in "two meanings"). The concept of ambiguity is generally contrasted with vagueness. In ambiguity, specific and distinct interpretations are permitted (although some may not be immediately obvious), whereas with vague information it is difficult to form any interpretation at the desired level of specificity. Linguistic forms Lexical ambiguity is contrasted with semantic ambiguity. The former represents a choice between a finite number of known and meaningful context-dependent interpretations. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |