|
Gyrase
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. The enzyme causes negative supercoiling of the DNA or relaxes positive supercoils. It does so by looping the template so as to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite '' Plasmodium falciparum'' and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
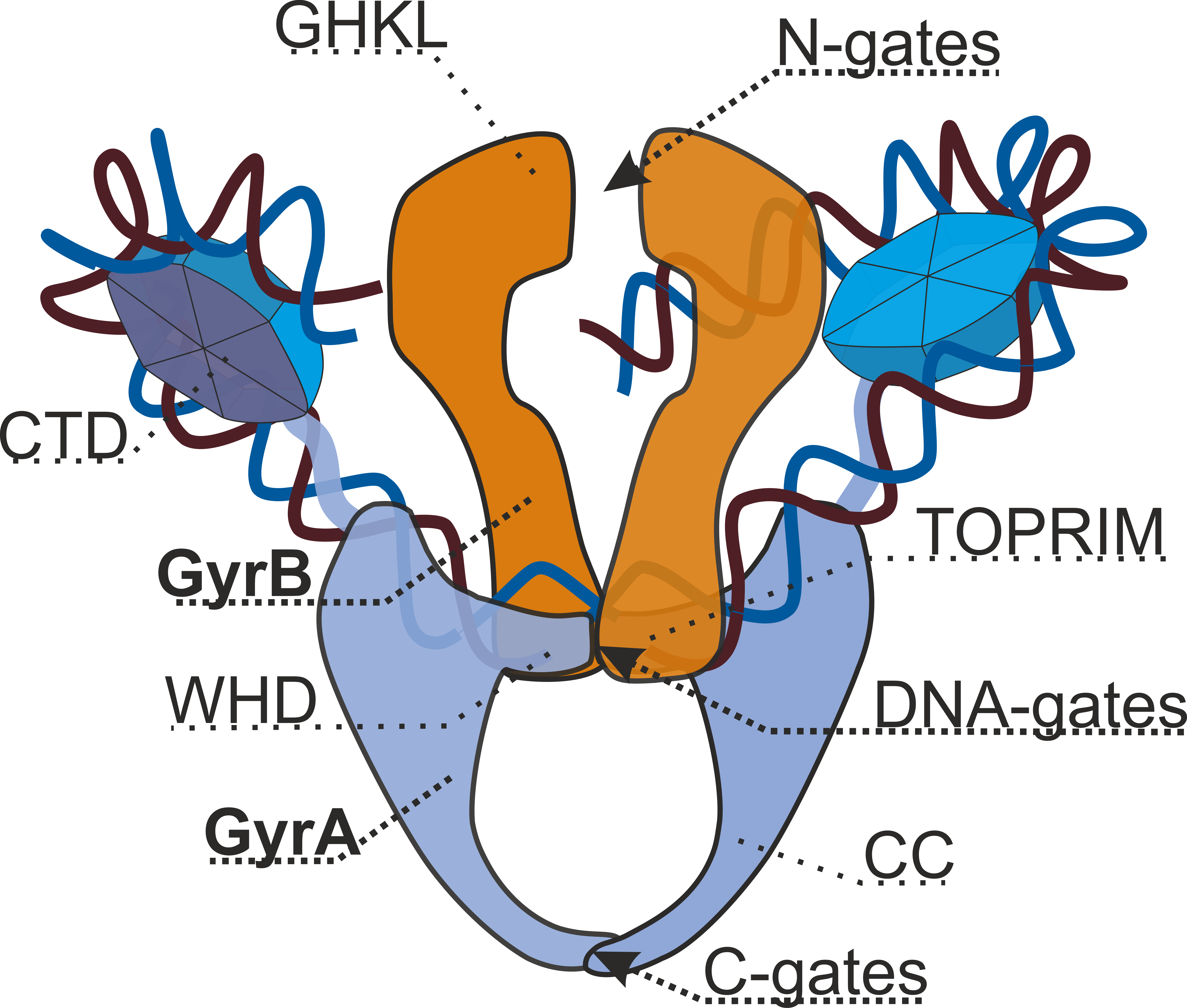
Gyrase Structure Dmitry Sutormin Eng
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. The enzyme causes negative supercoiling of the DNA or relaxes positive supercoils. It does so by looping the template so as to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite '' Plasmodium falciparum'' and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
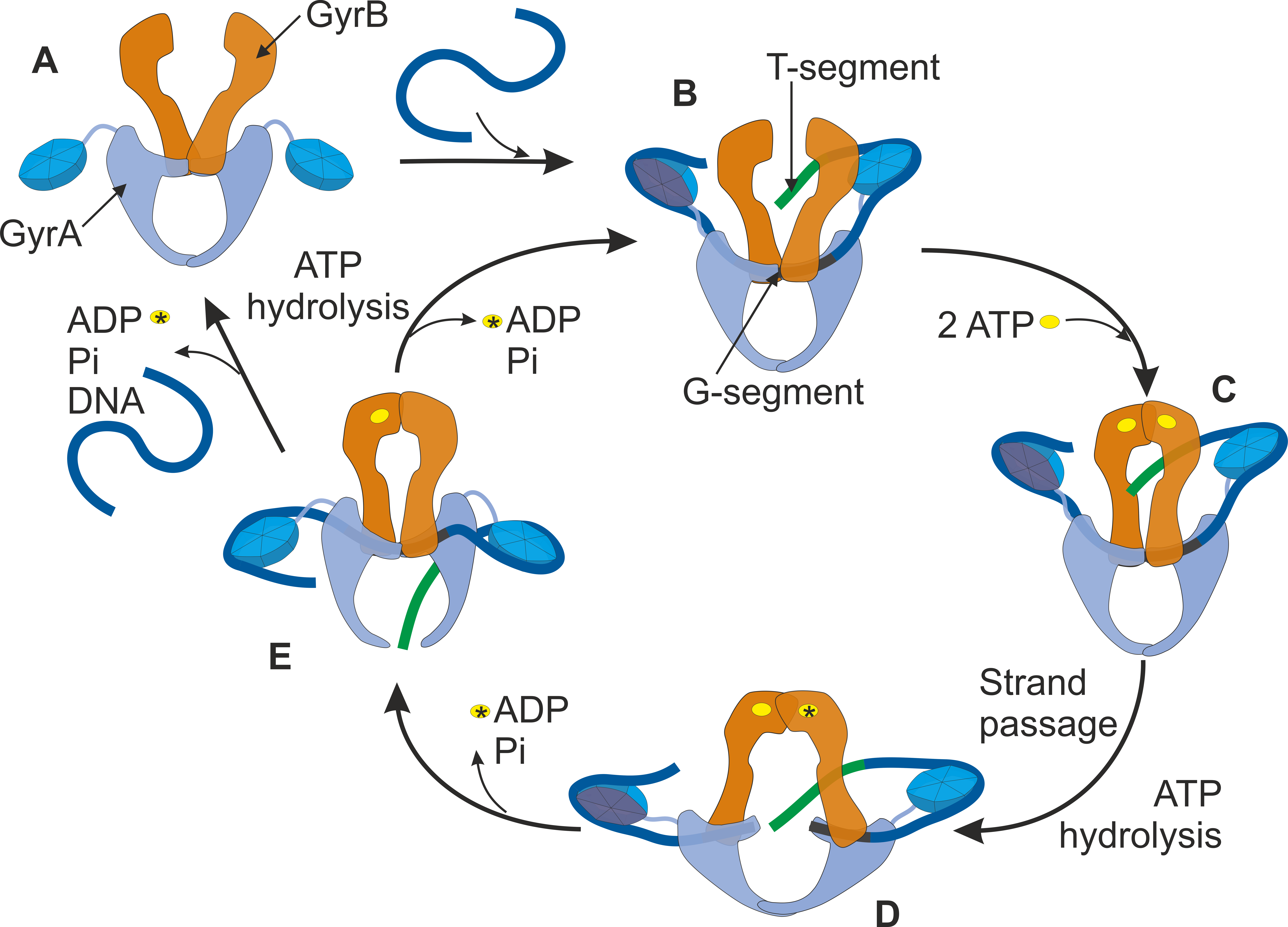
Gyrase Catalytic Cycle Eng Sutormin Eng
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. The enzyme causes negative supercoiling of the DNA or relaxes positive supercoils. It does so by looping the template so as to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite ''Plasmodium falciparum'' and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiotics, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Type II Topoisomerase
Type II topoisomerases are topoisomerases that cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change the linking number of circular DNA by ±2. Topoisomerases are ubiquitous enzymes, found in all living organisms. In animals, topoisomerase II is a chemotherapy target. In prokaryotes, gyrase is an antibacterial target. Indeed, these enzymes are of interest for a wide range of effects. Function Type II topoisomerases increase or decrease the linking number of a DNA loop by 2 units, and it promotes chromosome disentanglement. For example, DNA gyrase, a type II topoisomerase observed in '' E. coli'' and most other prokaryotes, introduces negative supercoils and decreases the linking number by 2. Gyrase is also able to remove knots from the bacterial chromosome. Along with gyrase, most prokaryotes also contain a second type IIA topoisomerase, t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Topoisomerase
DNA topoisomerases (or topoisomerases) are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA. Topological issues in DNA arise due to the intertwined nature of its double-helical structure, which, for example, can lead to overwinding of the DNA duplex during DNA replication and transcription. If left unchanged, this torsion would eventually stop the DNA or RNA polymerases involved in these processes from continuing along the DNA helix. A second topological challenge results from the linking or tangling of DNA during replication. Left unresolved, links between replicated DNA will impede cell division. The DNA topoisomerases prevent and correct these types of topological problems. They do this by binding to DNA and cutting the sugar-phosphate backbone of either one (type I topoisomerases) or both (type II topoisomerases) of the DNA strands. This transien ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aminocoumarin
Aminocoumarin is a class of antibiotics that act by an inhibition of the DNA gyrase enzyme involved in the cell division in bacteria. They are derived from ''Streptomyces '' species, whose best-known representative – ''Streptomyces coelicolor'' – was completely sequenced in 2002. The aminocoumarin antibiotics include: * Novobiocin, Albamycin (Pharmacia And Upjohn) * Coumermycin * Clorobiocin Structure The core of aminocoumarin antibiotics is made up of a 3-amino-4,7-dihydroxycumarin ring, which is linked, e.g., with a sugar in 7-Position and a benzoic acid derivative in 3-Position. Clorobiocin is a natural antibiotic isolated from several ''Streptomyces'' strains and differs from novobiocin in that the methyl group at the 8 position in the coumarin ring of novobiocin is replaced by a chlorine atom, and the carbamoyl at the 3' position of the noviose sugar is substituted by a 5-methyl-2-pyrrolylcarbonyl group. Mechanism of action The aminocoumarin antibiotics are known i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Novobiocin
Novobiocin, also known as albamycin or cathomycin, is an aminocoumarin antibiotic that is produced by the actinomycete '' Streptomyces niveus'', which has recently been identified as a subjective synonym for ''S. spheroides'' a member of the class Actinomycetia. Other aminocoumarin antibiotics include clorobiocin and coumermycin A1.Alessandra da Silva Eustáquio (2004) Biosynthesis of aminocoumarin antibiotics in Streptomyces: Generation of structural analogues by genetic engineering and insights into the regulation of antibiotic production. DISSERTATION Novobiocin was first reported in the mid-1950s (then called streptonivicin). Clinical use It is active against ''Staphylococcus epidermidis'' and may be used to differentiate it from the other coagulase-negative ''Staphylococcus saprophyticus'', which is resistant to novobiocin, in culture. Novobiocin was licensed for clinical use under the tradename Albamycin (Upjohn) in the 1960s. Its efficacy has been demonstrated in preclini ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Topoisomerase IV
Topoisomerase IV is one of two Type II topoisomerases in bacteria, the other being DNA gyrase. Like gyrase, topoisomerase IV is able to pass one double-strand of DNA through another double-strand of DNA, thereby changing the linking number of DNA by two in each enzymatic step. Both share a hetero-4-mer structure formed by a symmetric homodimer of A/B heterodimers, usually named ''ParC'' and ''ParE''. Functions Topoisomerase IV has two functions in the cell. * First, it is responsible for unlinking, or decatenating, DNA following DNA replication. The double-helical nature of DNA and its semiconservative mode of replication causes the two newly replicated DNA strands to be interlinked. These links must be removed in order for the chromosome (and plasmids) to segregate into daughter cells so that cell division can complete. * Topoisomerase IV's second function in the cell is to relax positive supercoils. It shares this role with DNA gyrase, which is also able to relax positive super ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential. DNA is made up of a double helix of two complementary strands. The double helix describes the appearance of a double-stranded DNA which is thus composed of two linear strands that run opposite to each other and twist together to form. During replication, these strands are separated. Each strand of the original DNA molecule then serves as a template for the production of its counterpart, a process referred to as semiconservative replication. As a result of semi-conservative rep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nalidixic Acid
Nalidixic acid (tradenames Nevigramon, NegGram, Wintomylon and WIN 18,320) is the first of the synthetic quinolone antibiotics. In a technical sense, it is a naphthyridone, not a quinolone: its ring structure is a 1,8-naphthyridine nucleus that contains two nitrogen atoms, unlike quinoline, which has a single nitrogen atom. Synthetic quinolone antibiotics were discovered by George Lesher and coworkers as a byproduct of chloroquine manufacture in the 1960s; nalidixic acid itself was used clinically, starting in 1967. Nalidixic acid is effective primarily against Gram-negative bacteria, with minor anti-Gram-positive activity. In lower concentrations, it acts in a bacteriostatic manner; that is, it inhibits growth and reproduction. In higher concentrations, it is bactericidal, meaning that it kills bacteria instead of merely inhibiting their growth. It has historically been used for treating urinary tract infections, caused, for example, by ''Escherichia coli'', ''Proteus'', ''Sh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ciprofloxacin
Ciprofloxacin is a fluoroquinolone antibiotic used to treat a number of bacterial infections. This includes bone and joint infections, intra abdominal infections, certain types of infectious diarrhea, respiratory tract infections, skin infections, typhoid fever, and urinary tract infections, among others. For some infections it is used in addition to other antibiotics. It can be taken by mouth, as eye drops, as ear drops, or intravenously. Common side effects include nausea, vomiting, and diarrhea. Severe side effects include an increased risk of tendon rupture, hallucinations, and nerve damage. In people with myasthenia gravis, there is worsening muscle weakness. Rates of side effects appear to be higher than some groups of antibiotics such as cephalosporins but lower than others such as clindamycin. Studies in other animals raise concerns regarding use in pregnancy. No problems were identified, however, in the children of a small number of women who took the medication. It ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Supercoil
DNA supercoiling refers to the amount of twist in a particular DNA strand, which determines the amount of strain on it. A given strand may be "positively supercoiled" or "negatively supercoiled" (more or less tightly wound). The amount of a strand’s supercoiling affects a number of biological processes, such as compacting DNA and regulating access to the genetic code (which strongly affects DNA metabolism and possibly gene expression). Certain enzymes, such as topoisomerases, change the amount of DNA supercoiling to facilitate functions such as DNA replication and transcription. The amount of supercoiling in a given strand is described by a mathematical formula that compares it to a reference state known as "relaxed B-form" DNA. Overview In a "relaxed" double-helical segment of B-DNA, the two strands twist around the helical axis once every 10.4–10.5 base pairs of sequence. Adding or subtracting twists, as some enzymes do, imposes strain. If a DNA segment under twist s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriophage Mu
Bacteriophage Mu, also known as mu phage or mu bacteriophage, is a muvirus (the first of its kind to be identified) of the family ''Myoviridae'' which has been shown to cause genetic transposition. It is of particular importance as its discovery in ''Escherichia coli'' by Larry Taylor was among the first observations of insertion elements in a genome. This discovery opened up the world to an investigation of transposable elements and their effects on a wide variety of organisms. While Mu was specifically involved in several distinct areas of research (including ''E. coli'', maize, and HIV), the wider implications of transposition and insertion transformed the entire field of genetics. Anatomy Phage Mu is nonenveloped, with a head and a tail. The head has an icosahedral structure of about 54 nm in width. The neck is knob-like, and the tail is contractile with a base plate and six short terminal fibers. The genome has been fully sequenced and consists of 36,717 nucleotides, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |