|
Genealogical DNA Test
A genealogical DNA test is a DNA-based Genetic testing, genetic test used in genetic genealogy that looks at specific locations of a person's genome in order to find or verify ancestral genealogical relationships, or (with lower reliability) to estimate the ethnicity, ethnic mixture of an individual. Since different testing companies use different ethnic reference groups and different matching algorithms, ethnicity estimates for an individual vary between tests, sometimes dramatically. Three principal types of genealogical DNA tests are available, with each looking at a different part of the genome and being useful for different types of genealogical research: Genealogical DNA test#Autosomal DNA (atDNA) testing, autosomal (atDNA), Genealogical DNA test#Mitochondrial DNA (mtDNA) testing, mitochondrial (mtDNA), and Genealogical DNA test#Y-chromosome (Y-DNA) testing, Y-chromosome (Y-DNA). Autosomal tests may result in a large number of DNA matches to both males and females who have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Testing
Genetic testing, also known as DNA testing, is used to identify changes in DNA sequence or chromosome structure. Genetic testing can also include measuring the results of genetic changes, such as RNA analysis as an output of gene expression, or through biochemical analysis to measure specific protein output. In a medical setting, genetic testing can be used to diagnose or rule out suspected genetic disorders, predict risks for specific conditions, or gain information that can be used to customize medical treatments based on an individual's genetic makeup. Genetic testing can also be used to determine biological relatives, such as a child's biological parentage (genetic mother and father) through DNA paternity testing, or be used to broadly predict an individual's ancestry. Genetic testing of plants and animals can be used for similar reasons as in humans (e.g. to assess relatedness/ancestry or predict/diagnose genetic disorders), to gain information used for selective breed ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
23andMe
23andMe Holding Co. is an American personal genomics and biotechnology company based in South San Francisco, California. It is best known for providing a direct-to-consumer genetic testing service in which customers provide a saliva testing, saliva sample that is laboratory analysed, using SNP genotyping, single nucleotide polymorphism genotyping, to generate reports relating to the customer's ancestry and genetic predispositions to health-related topics. The company's name is derived from the 23 pairs of chromosomes in a diploid human cell (biology), cell. Founded in 2006, 23andMe soon became the first company to begin offering autosomal DNA testing for ancestry, which all other major companies now use. Its saliva-based direct-to-consumer genetic testing business was named "Invention of the Year" by ''Time (magazine), Time'' in 2008. The company had a previously fraught relationship with the United States Food and Drug Administration (FDA) due to its genetic health tests; as of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hyper Variable Region
A hypervariable region (HVR) is a location within a sequence where polymorphisms frequently occur. It is used in two contexts: * In the case of nucleic acids, an HVR is where base pairs frequently change. This can be due to a change in the number of repeats (which is seen in eukaryotic nuclear DNA) or simply low selective pressure allowing a great number of substitutions and indels (as in the case of mitochondrial DNA D-loop and 16S rRNA). * In the case of antibodies, an HVR is where most of the differences among antibodies occur. This region is also called the complementarity-determining region. Because there already is a separate article for the antibody region, this article will focus on the nucleic acid case. Mitochondrial There are two mitochondrial hypervariable regions used in human mitochondrial genealogical DNA testing. HVR1 is considered a "low resolution" region and HVR2 is considered a "high resolution" region. Getting HVR1 and HVR2 DNA tests can help determine one ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MtDNA
Mitochondrial DNA (mtDNA and mDNA) is the DNA located in the mitochondria organelles in a eukaryotic cell that converts chemical energy from food into adenosine triphosphate (ATP). Mitochondrial DNA is a small portion of the DNA contained in a eukaryotic cell; most of the DNA is in the cell nucleus, and, in plants and algae, the DNA also is found in plastids, such as chloroplasts. Mitochondrial DNA is responsible for coding of 13 essential subunits of the complex oxidative phosphorylation (OXPHOS) system which has a role in cellular energy conversion. Human mitochondrial DNA was the first significant part of the human genome to be sequenced. This sequencing revealed that human mtDNA has 16,569 base pairs and encodes 13 proteins. As in other vertebrates, the human mitochondrial genetic code differs slightly from nuclear DNA. Since animal mtDNA evolves faster than nuclear genetic markers, it represents a mainstay of phylogenetics and evolutionary biology. It also permits tra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
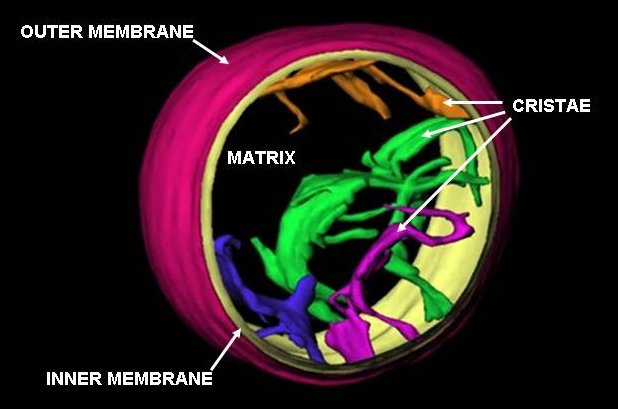
Mitochondrion
A mitochondrion () is an organelle found in the cell (biology), cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double lipid bilayer, membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'', meaning a thread-like granule, was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase popularized by Philip Siekevitz in a 1957 ''Scientific American'' article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). The multicellular animal ''Henneguya zschokkei, Henneguya salminicola'' is known to have retained mitochondrion-related organelles despite a complete loss of their mitochondrial genome. A large number ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Centimorgan
In genetics, a centimorgan (abbreviated cM) or map unit (m.u.) is a unit for measuring genetic linkage. It is defined as the distance between chromosome positions (also termed loci or markers) for which the expected average number of intervening chromosomal crossovers in a single generation is 0.01. It is often used to infer distance along a chromosome. However, it is not a true physical distance. Relation to physical distance The number of base pairs to which it corresponds varies widely across the genome (different regions of a chromosome have different propensities towards crossover) and it also depends on whether the meiosis in which the crossing-over takes place is a part of oogenesis (formation of female gametes) or spermatogenesis (formation of male gametes). In humans one centimorgan corresponds to about 1 Mb (1,000,000 base pairs or nucleotides) on average. The relationship is only rough, as the physical chromosomal distance corresponding to one centimorgan varies f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Illumina (company)
Illumina, Inc. is an American biotechnology company, headquartered in San Diego, California. Incorporated on April 1, 1998, Illumina develops, manufactures, and markets integrated systems for the analysis of genetic variation and biological function. The company provides a line of products and services that serves the sequencing, genotyping and gene expression, and proteomics markets, and serves more than 155 countries. Illumina's customers include genomic research centers, pharmaceutical companies, academic institutions, clinical research organizations, and biotechnology companies. History Illumina was founded in April 1998 by David Walt, Larry Bock, John Stuelpnagel, Anthony Czarnik, and Mark Chee. While working with CW Group, a venture-capital firm, Bock and Stuelpnagel uncovered what would become Illumina's BeadArray technology at Tufts University and negotiated an exclusive license to that technology. In 1999, Illumina acquired Spyder Instruments (founded by Michal Le ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, a Guanine, G nucleotide present at a specific location in a reference genome may be replaced by an Adenine, A in a minority of individuals. The two possible nucleotide variations of this SNP – G or A – are called alleles. SNPs can help explain differences in susceptibility to a wide range of diseases across a population. For example, a common SNP in the Factor H, CFH gene is associated with increased risk of age-related macular degeneration. Differences in the severity of an illness or response to treatments may also be manifestations of genetic variations caused by SNPs. For example, two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autosome
An autosome is any chromosome that is not a sex chromosome. The members of an autosome pair in a diploid cell have the same morphology, unlike those in allosomal (sex chromosome) pairs, which may have different structures. The DNA in autosomes is collectively known as atDNA or auDNA. For example, humans have a diploid genome that usually contains 22 pairs of autosomes and one allosome pair (46 chromosomes total). The autosome pairs are labeled with numbers (1–22 in humans) roughly in order of their sizes in base pairs, while allosomes are labelled with their letters. By contrast, the allosome pair consists of two X chromosomes in females or one X and one Y chromosome in males. Unusual combinations XYY, XXY, XXX, XXXX, XXXXX or XXYY, among other irregular combinations, are known to occur and usually cause developmental abnormalities. Autosomes still contain sexual determination genes even though they are not sex chromosomes. For example, the SRY gene on the Y chromos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Microarray
A DNA microarray (also commonly known as a DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10−12 moles) of a specific DNA sequence, known as '' probes'' (or ''reporters'' or '' oligos''). These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA (also called anti-sense RNA) sample (called ''target'') under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |