|
Frameshift
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure. It has been described mainly in viruses (especially retroviruses), retrotransposons and bacterial insertion elements, and also in some cellular genes. Process overview Proteins are translated by reading tri-nucleotides on the mRNA strand, also known as codons, from one end of the mRNA to the other (from the 5' to the 3' end) starting with the amino acid methionine as the start (initiation) codon AUG. Each codon is translated into a single amino acid. The code itself is considered degenerate, meaning that a particular amino acid can be specified by more than one codons. However, a shift of any number of nucleo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Slippery Sequence
A slippery sequence is a small section of codon nucleotide sequences (usually UUUAAAC) that controls the rate and chance of ribosomal frameshifting. A slippery sequence causes a faster ribosomal transfer which in turn can cause the reading ribosome to "slip." This allows a tRNA to shift by 1 base (−1) after it has paired with its anticodon, changing the reading frame. A −1 frameshift triggered by such a sequence is a Programmed −1 Ribosomal Frameshift. It is followed by a spacer region, and an RNA secondary structure. Such sequences are common in virus polyproteins. The frameshift occurs due to wobble pairing. The Gibbs free energy of secondary structures downstream give a hint at how often frameshift happens. Tension on the mRNA molecule also plays a role. A list of slippery sequences found in animal viruses is available from Huang et al. Slippery sequences that cause a 2-base slip (−2 frameshift) have been constructed out of the HIV UUUUUUA sequence. See also *Nucl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Slippery Sequence
A slippery sequence is a small section of codon nucleotide sequences (usually UUUAAAC) that controls the rate and chance of ribosomal frameshifting. A slippery sequence causes a faster ribosomal transfer which in turn can cause the reading ribosome to "slip." This allows a tRNA to shift by 1 base (−1) after it has paired with its anticodon, changing the reading frame. A −1 frameshift triggered by such a sequence is a Programmed −1 Ribosomal Frameshift. It is followed by a spacer region, and an RNA secondary structure. Such sequences are common in virus polyproteins. The frameshift occurs due to wobble pairing. The Gibbs free energy of secondary structures downstream give a hint at how often frameshift happens. Tension on the mRNA molecule also plays a role. A list of slippery sequences found in animal viruses is available from Huang et al. Slippery sequences that cause a 2-base slip (−2 frameshift) have been constructed out of the HIV UUUUUUA sequence. See also *Nucl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Codons
The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries. The codons specify which amino acid will be added next during protein biosynthesis. With some exceptions, a three-nucleotide codon in a nucleic acid sequence specifies a single amino acid. The vast majority of genes are encoded with a single scheme (see the RNA codon table). That scheme is often referred to as the canonical or standard genetic code, or simply ''the'' genetic code, though variant codes (such as in mitochondria) exist. History Effo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Vertebrate Mitochondrial Code
The vertebrate mitochondrial code (translation table 2) is the genetic code found in the mitochondria of all vertebrata. Evolution AGA and AGG were thought to have become mitochondrial stop codons early in vertebrate evolution. However, at least in humans it has now been shown that AGA and AGG sequences are not recognized as termination codons. A -1 Mitochondrial ribosome, mitoribosome frameshift occurs at the AGA and AGG codons predicted to terminate the CO1 and ND6 open reading frames (ORFs), and consequently both ORFs terminate in the standard UAG codon. Incomplete stop codons Mitochondrial genes in some vertebrates (including humans) have incomplete stop codons ending in U or UA, which become complete termination codons (UAA) upon subsequent polyadenylation. Translation table : The codon AUG both codes for methionine and serves as an initiation site: the first AUG in an mRNA's coding region is where translation into protein begins. Differences from the standard code ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MT-ATP6
''MT-ATP6'' (or ''ATP6'') is a mitochondrial gene with the full name 'mitochondrially encoded ATP synthase membrane subunit 6' that encodes the ATP synthase Fo subunit 6 (or subunit/chain A). This subunit belongs to the Fo complex of the large, transmembrane F-type ATP synthase. This enzyme, which is also known as complex V, is responsible for the final step of oxidative phosphorylation in the electron transport chain. Specifically, one segment of ATP synthase allows positively charged ions, called protons, to flow across a specialized membrane inside mitochondria. Another segment of the enzyme uses the energy created by this proton flow to convert a molecule called adenosine diphosphate (ADP) to ATP. Mutations in the ''MT-ATP6'' gene have been found in approximately 10 to 20 percent of people with Leigh syndrome. Structure The ''MT-ATP6'' gene provides information for making a protein that is essential for normal mitochondrial function. The human ''MT-ATP6'' gene, located ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MT-ATP8
''MT-ATP8'' (or ''ATP8'') is a mitochondrial gene with the full name 'mitochondrially encoded ATP synthase membrane subunit 8' that encodes a subunit of mitochondrial ATP synthase, ATP synthase Fo subunit 8 (or subunit A6L). This subunit belongs to the Fo complex of the large, transmembrane F-type ATP synthase. This enzyme, which is also known as complex V, is responsible for the final step of oxidative phosphorylation in the electron transport chain. Specifically, one segment of ATP synthase allows positively charged ions, called protons, to flow across a specialized membrane inside mitochondria. Another segment of the enzyme uses the energy created by this proton flow to convert a molecule called adenosine diphosphate (ADP) to ATP. Subunit 8 differs in sequence between Metazoa, plants and Fungi. Structure The ATP synthase protein 8 of human and other mammals is encoded in the mitochondrial genome by the ''MT-ATP8'' gene. When the complete human mitochondrial genome was ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
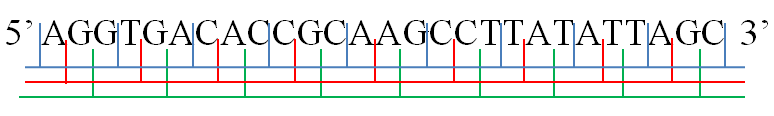
Reading Frame
In molecular biology, a reading frame is a way of dividing the sequence of nucleotides in a nucleic acid ( DNA or RNA) molecule into a set of consecutive, non-overlapping triplets. Where these triplets equate to amino acids or stop signals during translation, they are called codons. A single strand of a nucleic acid molecule has a phosphoryl end, called the 5′-end, and a hydroxyl or 3′-end. These define the 5′→3′ direction. There are three reading frames that can be read in this 5′→3′ direction, each beginning from a different nucleotide in a triplet. In a double stranded nucleic acid, an additional three reading frames may be read from the other, complementary strand in the 5′→3′ direction along this strand. As the two strands of a double-stranded nucleic acid molecule are antiparallel, the 5′→3′ direction on the second strand corresponds to the 3′→5′ direction along the first strand. In general, at the most, one reading frame in a given se ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
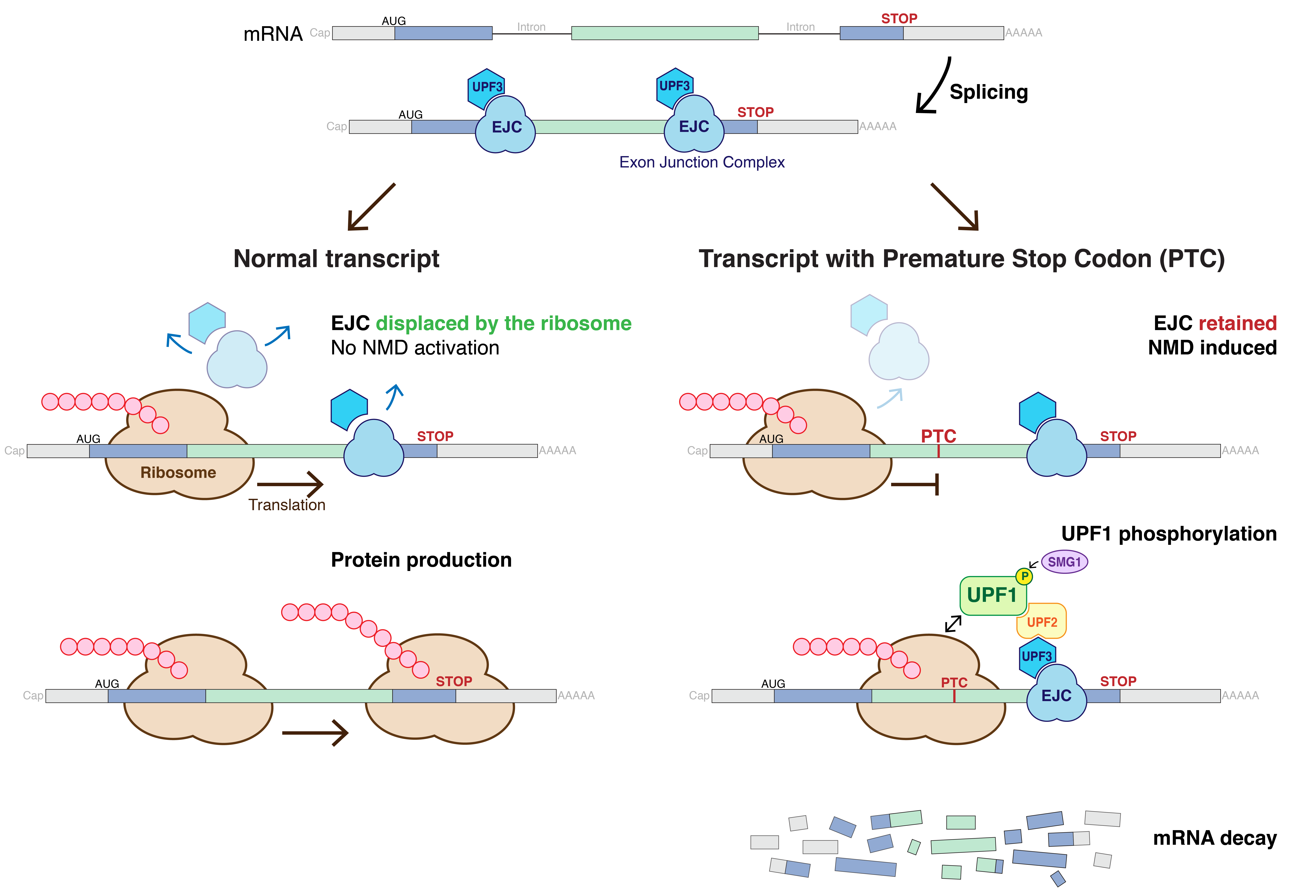
Nonsense-mediated Decay
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Translation of these aberrant mRNAs could, in some cases, lead to deleterious gain-of-function or dominant-negative activity of the resulting proteins. NMD was first described in human cells and in yeast almost simultaneously in 1979. This suggested broad phylogenetic conservation and an important biological role of this intriguing mechanism. NMD was discovered when it was realized that cells often contain unexpectedly low concentrations of mRNAs that are transcribed from alleles carrying nonsense mutations. Nonsense mutations code for a premature stop codon which causes the protein to be shortened. The truncated protein may or may not be functional, depending on the severity of what is not translated. In human genetics, NMD has the possibility to not only limit the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
+1 Translational Frameshift Mechanism
A +1 (pronounced "plus one") is a person who accompanies someone to an event. The term may also refer to: Arts, entertainment, and media * ''+1'' (album), 2008, by Kaela Kimura * ''+1'' (film) (also known as ''Plus One''), 2013 * "+1" (song), by French DJ Martin Solveig * +1 Records, an extension of the music management, publicity & marketing company +1 Music * +1 (also known as ''Plus One''), a "lifeline" in the ''Who Wants to Be a Millionaire?'' franchise Technology * +1 button, the "like" button implemented by Google applications * +1, the country calling code (telephone calling prefix) for countries in the North American Dialing Plan, for the list of codes, see list of North American Numbering Plan area codes * +1, a suffix attached to the name of a television timeshift channel See also * 1-up * And 1 (other) * Hear, hear * Infinity + 1 * Me Too (other) * One (other) * OnePlus * Plus One (other) * Successor function In mathe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Influenza
Influenza, commonly known as "the flu", is an infectious disease caused by influenza viruses. Symptoms range from mild to severe and often include fever, runny nose, sore throat, muscle pain, headache, coughing, and fatigue. These symptoms begin from one to four days after exposure to the virus (typically two days) and last for about 2–8 days. Diarrhea and vomiting can occur, particularly in children. Influenza may progress to pneumonia, which can be caused by the virus or by a subsequent bacterial infection. Other complications of infection include acute respiratory distress syndrome, meningitis, encephalitis, and worsening of pre-existing health problems such as asthma and cardiovascular disease. There are four types of influenza virus, termed influenza viruses A, B, C, and D. Aquatic birds are the primary source of Influenza A virus (IAV), which is also widespread in various mammals, including humans and pigs. Influenza B virus (IBV) and Influenza C virus (ICV) pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Wobble Base Pair
A wobble base pair is a pairing between two nucleotides in RNA molecules that does not follow Watson-Crick base pair rules. The four main wobble base pairs are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A), and hypoxanthine-cytosine (I-C). In order to maintain consistency of nucleic acid nomenclature, "I" is used for hypoxanthine because hypoxanthine is the nucleobase of inosine; nomenclature otherwise follows the names of nucleobases and their corresponding nucleosides (e.g., "G" for both guanine and guanosine – as well as for deoxyguanosine). The thermodynamic stability of a wobble base pair is comparable to that of a Watson-Crick base pair. Wobble base pairs are fundamental in RNA secondary structure and are critical for the proper translation of the genetic code. Brief history In the genetic code, there are 43 = 64 possible codons (3 nucleotide sequences). For translation, each of these codons requires a tRNA molecule with an anticodon wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |