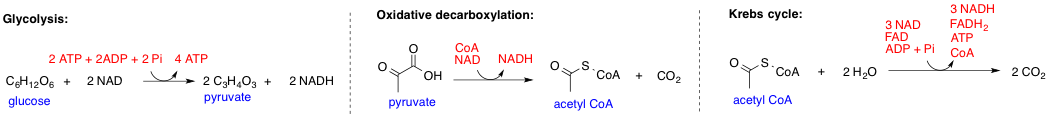|
Enzyme Tunnel
Metabolite channeling is the passing of the intermediary metabolic product of one enzyme directly to another enzyme or active site without its release into solution. When several consecutive enzymes of a metabolic pathway channel substrates between themselves, this is called a metabolon. Channeling can make a metabolic pathway more rapid and efficient than it would be if the enzymes were randomly distributed in the cytosol, or prevent the release of unstable intermediates. It can also protect an intermediate from being consumed by competing reactions catalyzed by other enzymes. Mechanisms for channeling Channeling can occur in several ways. One possibility, which occurs in the pyruvate dehydrogenase complex, is by a substrate being attached to a flexible arm that moves between several active sites (not very likely). Another possibility is by two active sites being connected by a tunnel through the protein and the substrate moving through the tunnel; this is seen in tryptophan synth ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) and residues that catalyse a reaction of that substrate (catalytic site). Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their function. The active si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metabolon
In biochemistry, a metabolon is a temporary structural-functional complex formed between sequential enzymes of a metabolic pathway, held together both by non-covalent interactions and by structural elements of the cell, such as integral membrane proteins and proteins of the cytoskeleton. The formation of metabolons allows the intermediate product from one enzyme to be passed (channelling) directly into the active site of the next consecutive enzyme of the metabolic pathway. The citric acid cycle is an example of a metabolon that facilitates substrate channeling. Another example is the dhurrin synthesis pathway in sorghum, in which the enzymes assemble as a metabolon in lipid membranes. During the functioning of metabolons, the amount of water needed to hydrate the enzymes is reduced and enzyme activity is increased. History The concept of structural-metabolic cellular complexes was first conceived in 1970 by A. M. Kuzin of the USSR Academy of Sciences, and adopted in 1972 by Pau ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metabolic Pathway
In biochemistry, a metabolic pathway is a linked series of chemical reactions occurring within a cell. The reactants, products, and intermediates of an enzymatic reaction are known as metabolites, which are modified by a sequence of chemical reactions catalyzed by enzymes. In most cases of a metabolic pathway, the product of one enzyme acts as the substrate for the next. However, side products are considered waste and removed from the cell. These enzymes often require dietary minerals, vitamins, and other cofactors to function. Different metabolic pathways function based on the position within a eukaryotic cell and the significance of the pathway in the given compartment of the cell. For instance, the, electron transport chain, and oxidative phosphorylation all take place in the mitochondrial membrane. In contrast, glycolysis, pentose phosphate pathway, and fatty acid biosynthesis all occur in the cytosol of a cell. There are two types of metabolic pathways that are character ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytosol
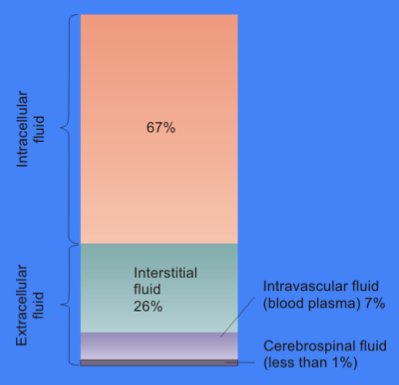
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells (intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into many compartments. In the eukaryotic cell, the cytosol is surrounded by the cell membrane and is part of the cytoplasm, which also comprises the mitochondria, plastids, and other organelles (but not their internal fluids and structures); the cell nucleus is separate. The cytosol is thus a liquid matrix around the organelles. In prokaryotes, most of the chemical reactions of metabolism take place in the cytosol, while a few take place in membranes or in the periplasmic space. In eukaryotes, while many metabolic pathways still occur in the cytosol, others take place within organelles. The cytosol is a complex mixture of substances dissolved in water. Although water forms the large majority of the cytosol, its structure and prope ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyruvate Dehydrogenase Complex
Pyruvate dehydrogenase complex (PDC) is a complex of three enzymes that converts pyruvate into acetyl-CoA by a process called pyruvate decarboxylation. Acetyl-CoA may then be used in the citric acid cycle to carry out cellular respiration, and this complex links the glycolysis metabolic pathway to the citric acid cycle. Pyruvate decarboxylation is also known as the "pyruvate dehydrogenase reaction" because it also involves the oxidation of pyruvate. This multi-enzyme complex is related structurally and functionally to the oxoglutarate dehydrogenase and branched-chain oxo-acid dehydrogenase multi-enzyme complexes. Reaction The reaction catalysed by pyruvate dehydrogenase complex is: Structure Pyruvate dehydrogenase (E1) The E1 subunit, called the pyruvate dehydrogenase subunit, has a structure that consists of two chains (an “ɑ” and “ꞵ” chain). A magnesium ion forms a 4-coordinate complex with three, polar amino acid residues (Asp, Asn, and Tyr) located on t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tryptophan Synthase
Tryptophan synthase or tryptophan synthetase is an enzyme () that catalyses the final two steps in the biosynthesis of tryptophan. It is commonly found in Eubacteria, Archaebacteria, Protista, Fungi, and Plantae. However, it is absent from Animalia. It is typically found as an α2β2 tetramer. The α subunits catalyze the reversible formation of indole and glyceraldehyde-3-phosphate (G3P) from indole-3-glycerol phosphate (IGP). The β subunits catalyze the irreversible condensation of indole and serine to form tryptophan in a pyridoxal phosphate (PLP) dependent reaction. Each α active site is connected to a β active site by a 25 angstrom long hydrophobic channel contained within the enzyme. This facilitates the diffusion of indole formed at α active sites directly to β active sites in a process known as substrate channeling. The active sites of tryptophan synthase are allosterically coupled. Enzyme structure Subunits: Tryptophan synthase typically exists as an α-ββ-α comple ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dihydrofolate Reductase
Dihydrofolate reductase, or DHFR, is an enzyme that reduces dihydrofolic acid to tetrahydrofolic acid, using NADPH as an electron donor, which can be converted to the kinds of tetrahydrofolate cofactors used in 1-carbon transfer chemistry. In humans, the DHFR enzyme is encoded by the ''DHFR'' gene. It is found in the q11→q22 region of chromosome 5. Bacterial species possess distinct DHFR enzymes (based on their pattern of binding diaminoheterocyclic molecules), but mammalian DHFRs are highly similar. Structure A central eight-stranded beta-pleated sheet makes up the main feature of the polypeptide backbone folding of DHFR. Seven of these strands are parallel and the eighth runs antiparallel. Four alpha helices connect successive beta strands. Residues 9 – 24 are termed "Met20" or "loop 1" and, along with other loops, are part of the major subdomain that surround the active site. The active site is situated in the N-terminal half of the sequence, which includes a conse ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thymidylate Synthase
Thymidylate synthase (TS) () is an enzyme that catalyzes the conversion of deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP). Thymidine is one of the nucleotides in DNA. With inhibition of TS, an imbalance of deoxynucleotides and increased levels of dUMP arise. Both cause DNA damage. Function The following reaction is catalyzed by thymidylate synthase: : 5,10-methylenetetrahydrofolate + dUMP \rightleftharpoons dihydrofolate + dTMP By means of reductive methylation, deoxyuridine monophosphate (dUMP) and N5,N10-methylene tetrahydrofolate are together used to form dTMP, yielding dihydrofolate as a secondary product. This provides the sole de novo pathway for production of dTMP and is the only enzyme in folate metabolism in which the 5,10-methylenetetrahydrofolate is oxidised during one-carbon transfer. The enzyme is essential for regulating the balanced supply of the 4 DNA precursors in normal DNA replication: defects in the enzyme activity affectin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Herbert Gutfreund
Herbert Gutfreund (21 October 1921 – 21 March 2021), better known as Freddie Gutfreund, was a British biochemist of Austrian origin, and Emeritus Professor at the University of Bristol. Gutfreund died in March 2021 at the age of 99. Early life and education Gutfreund was born on 21 October 1921 in Vienna en, Viennese , iso_code = AT-9 , registration_plate = W , postal_code_type = Postal code , postal_code = , timezone = CET , utc_offset = +1 , timezone_DST ... to a middle-class professional family, the son of Clara (Pisko) and Paul Gutfreund. His father was a civil engineer, and on his mother's side there were several scientists including the physicist Karl Weissenberg. He had all his early education in Vienna. However, the political turmoil of the 1930s forced him to leave Austria for England after the Anschluss of 1938. He joined an agricultural training scheme and became an accom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |