|
Endosomal Sorting Complexes Required For Transport
The endosomal sorting complexes required for transport (ESCRT) machinery is made up of cytosolic protein complexes, known as ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III. Together with a number of accessory proteins, these ESCRT complexes enable a unique mode of membrane remodeling that results in membranes bending/budding away from the cytoplasm. These ESCRT components have been isolated and studied in a number of organisms including yeast and humans. A eukaryotic signature protein, the machinery is found in all eukaryotes and some archaea. The ESCRT machinery plays a vital role in a number of cellular processes including multivesicular body (MVB) biogenesis, cellular abscission, and viral budding. Multivesicular body (MVB) biogenesis is a process in which ubiquitin tagged proteins enter organelles called endosomes via the formation of vesicles. This process is essential for cells to destroy misfolded and damaged proteins. Without ESCRT machinery, these proteins can build up and lead ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endosome
Endosomes are a collection of intracellular sorting organelles in eukaryotic cells. They are parts of endocytic membrane transport pathway originating from the trans Golgi network. Molecules or ligands internalized from the plasma membrane can follow this pathway all the way to lysosomes for degradation or can be recycled back to the cell membrane in the endocytic cycle. Molecules are also transported to endosomes from the trans Golgi network and either continue to lysosomes or recycle back to the Golgi apparatus. Endosomes can be classified as early, sorting, or late depending on their stage post internalization. Endosomes represent a major sorting compartment of the endomembrane system in cells. Function Endosomes provide an environment for material to be sorted before it reaches the degradative lysosome. For example, low-density lipoprotein (LDL) is taken into the cell by binding to the LDL receptor at the cell surface. Upon reaching early endosomes, the LDL dissociates ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ESCRT Machinery-Corrected
The endosomal sorting complexes required for transport (ESCRT) machinery is made up of cytosolic protein complexes, known as ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III. Together with a number of accessory proteins, these ESCRT complexes enable a unique mode of membrane remodeling that results in membranes bending/budding away from the cytoplasm. These ESCRT components have been isolated and studied in a number of organisms including yeast and humans. A eukaryotic signature protein, the machinery is found in all eukaryotes and some archaea. The ESCRT machinery plays a vital role in a number of cellular processes including multivesicular body (MVB) biogenesis, cellular abscission, and viral budding. Multivesicular body (MVB) biogenesis is a process in which ubiquitin tagged proteins enter organelles called endosomes via the formation of vesicles. This process is essential for cells to destroy misfolded and damaged proteins. Without ESCRT machinery, these proteins can build up and lead ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coiled-coil
A coiled coil is a structural motif in proteins in which 2–7 alpha helix, alpha-helices are coiled together like the strands of a rope. (Protein dimer, Dimers and Protein trimer, trimers are the most common types.) Many coiled coil-type proteins are involved in important biological functions, such as the regulation of gene expression — e.g., transcription factors. Notable examples are the oncoproteins c-Fos and c-Jun, as well as the muscle protein tropomyosin. Discovery The possibility of coiled coils for α-keratin was initially somewhat controversial. Linus Pauling and Francis Crick independently came to the conclusion that this was possible at about the same time. In the summer of 1952, Pauling visited the laboratory in England where Crick worked. Pauling and Crick met and spoke about various topics; at one point, Crick asked whether Pauling had considered "coiled coils" (Crick came up with the term), to which Pauling said he had. Upon returning to the United States, Paul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antiparallel (biochemistry)
In biochemistry, two biopolymers are antiparallel if they run parallel to each other but with opposite directionality (alignments). An example is the two complementary strands of a DNA double helix, which run in opposite directions alongside each other. Nucleic acids Nucleic acid molecules have a phosphoryl (5') end and a hydroxyl (3') end. This notation follows from organic chemistry nomenclature, and can be used to define the movement of enzymes such as DNA polymerases relative to the DNA strand in a non-arbitrary manner. G-quadruplexes G-quadruplexes, also known as G4 DNA are secondary structures found in nucleic acids that are rich in guanine. These structures are normally located at the telomeres (the ends of the chromosomes). The G-quadruplex can either be parallel or antiparallel depending on the loop configuration, which is a component of the structure. If all the DNA strands run in the same direction, it is termed to be a parallel quadruplex, and is known as a str ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dimer (chemistry)
A dimer () ('' di-'', "two" + ''-mer'', "parts") is an oligomer consisting of two monomers joined by bonds that can be either strong or weak, covalent or intermolecular. Dimers also have significant implications in polymer chemistry, inorganic chemistry, and biochemistry. The term ''homodimer'' is used when the two molecules are identical (e.g. A–A) and ''heterodimer'' when they are not (e.g. A–B). The reverse of dimerization is often called dissociation. When two oppositely charged ions associate into dimers, they are referred to as ''Bjerrum pairs'', after Niels Bjerrum. Noncovalent dimers Anhydrous carboxylic acids form dimers by hydrogen bonding of the acidic hydrogen and the carbonyl oxygen. For example, acetic acid forms a dimer in the gas phase, where the monomer units are held together by hydrogen bonds. Under special conditions, most OH-containing molecules form dimers, e.g. the water dimer. Excimers and exciplexes are excited structures with a short lifetime. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hse1
Heparanase, also known as HPSE, is an enzyme that acts both at the cell-surface and within the extracellular matrix to degrade polymeric heparan sulfate molecules into shorter chain length oligosaccharides. Synthesis and structure The protein is originally synthesised in an inactive 65 kDa proheparanase form in the golgi apparatus and transferred to late endosomes/lysosomes for transport to the cell-surface. In the lysosome it is proteolytically processed into its active form. Proteolytic processing results in the production of three products, * a linker peptide * an 8 kDa proheparanase fragment and * a 50 kDa proheparanase fragment The 8 kDa and 50 kDa fragments form a heterodimer and it is this heterodimer that constitutes the active heparanase molecule. The linker protein is so called because prior to its excision it physically links the 8 kDa and 50 kDa proheparanase fragments. Complete excision of the linker peptide appears to be a prerequisite to the complete activation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Vacuolar Protein Sorting
Vacuolar protein sorting proteins are involved in the intracellular sorting and delivery of soluble vacuolar A vacuole () is a membrane-bound organelle which is present in Plant cell, plant and Fungus, fungal Cell (biology), cells and some protist, animal, and bacterial cells. Vacuoles are essentially enclosed compartments which are filled with water ... proteins. References Single-pass transmembrane proteins {{genetics-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heterodimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", '' di-'' + '' -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins. A protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein-coupled cannabinoid receptors have the ability to form both homo- and heterodimers with several typ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
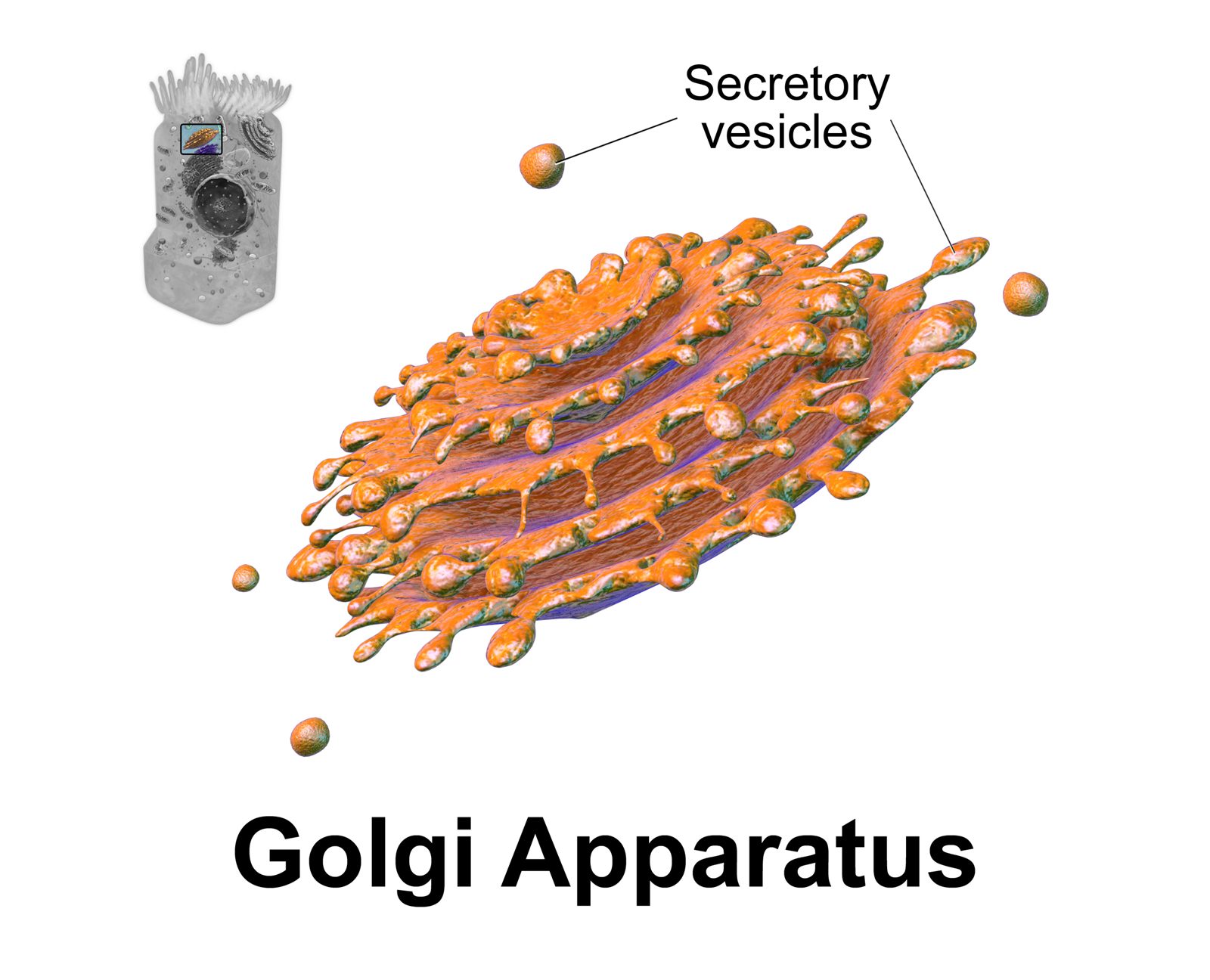
Golgi Apparatus
The Golgi apparatus (), also known as the Golgi complex, Golgi body, or simply the Golgi, is an organelle found in most eukaryotic cells. Part of the endomembrane system in the cytoplasm, it packages proteins into membrane-bound vesicles inside the cell before the vesicles are sent to their destination. It resides at the intersection of the secretory, lysosomal, and endocytic pathways. It is of particular importance in processing proteins for secretion, containing a set of glycosylation enzymes that attach various sugar monomers to proteins as the proteins move through the apparatus. It was identified in 1897 by the Italian scientist Camillo Golgi and was named after him in 1898. Discovery Owing to its large size and distinctive structure, the Golgi apparatus was one of the first organelles to be discovered and observed in detail. It was discovered in 1898 by Italian physician Camillo Golgi during an investigation of the nervous system. After first observing it under his ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysosome
A lysosome () is a membrane-bound organelle found in many animal cells. They are spherical vesicles that contain hydrolytic enzymes that can break down many kinds of biomolecules. A lysosome has a specific composition, of both its membrane proteins, and its lumenal proteins. The lumen's pH (~4.5–5.0) is optimal for the enzymes involved in hydrolysis, analogous to the activity of the stomach. Besides degradation of polymers, the lysosome is involved in various cell processes, including secretion, plasma membrane repair, apoptosis, cell signaling, and energy metabolism. Lysosomes act as the waste disposal system of the cell by digesting used materials in the cytoplasm, from both inside and outside the cell. Material from outside the cell is taken up through endocytosis, while material from the inside of the cell is digested through autophagy. The sizes of the organelles vary greatly—the larger ones can be more than 10 times the size of the smaller ones. They were discov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Subcellular Localization
The cells of eukaryotic organisms are elaborately subdivided into functionally-distinct membrane-bound compartments. Some major constituents of eukaryotic cells are: extracellular space, plasma membrane, cytoplasm, nucleus, mitochondria, Golgi apparatus, endoplasmic reticulum (ER), peroxisome, vacuoles, cytoskeleton, nucleoplasm, nucleolus, nuclear matrix and ribosomes. Bacteria also have subcellular localizations that can be separated when the cell is fractionated. The most common localizations referred to include the cytoplasm, the cytoplasmic membrane (also referred to as the inner membrane in Gram-negative bacteria), the cell wall (which is usually thicker in Gram-positive bacteria) and the extracellular environment. The cytoplasm, the cytoplasmic membrane and the cell wall are subcellular localizations, whereas the extracellular environment is clearly not. Most Gram-negative bacteria also contain an outer membrane and periplasmic space. Unlike eukaryotes, most bacteria cont ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |



_structure.png)