|
Endoglycosidase H
The enzyme endoglycosidase H () is an enzyme with systematic name ''glycopeptide-D-mannosyl-N4-(N-acetyl-D-glucosaminyl)2-asparagine 1,4-N-acetyl-beta-glucosaminohydrolase''. It is a highly specific endoglycosidase which cleaves asparagine-linked mannose rich oligosaccharides, but not highly processed complex oligosaccharides from glycoproteins. It is used for research purposes to deglycosylate glycoproteins and to monitor intracellular protein trafficking through the secretory pathway. Structure and activity Endoglycosidase H is isolated from '' Streptomyces plicatus'' or '' Streptomyces griseus''. Its molecular weight is 29 000 Daltons. The primary structure was described by Robbins et al. in 1984. Endoglycosidase H cleaves the bond in the diacetylchitobiose core of the oligosaccharide between two N-acetylglucosamine (GlcNAc) subunits directly proximal to the asparagine residue, generating a truncated sugar molecule with one N-acetylglucosamine residue remaining on the aspara ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Posttranslational Modification
Post-translational modification (PTM) is the covalent and generally enzymatic modification of proteins following protein biosynthesis. This process occurs in the endoplasmic reticulum and the golgi apparatus. Proteins are synthesized by ribosomes translating mRNA into polypeptide chains, which may then undergo PTM to form the mature protein product. PTMs are important components in cell signal transduction, signaling, as for example when prohormones are converted to hormones. Post-translational modifications can occur on the amino acid side chains or at the protein's C-terminus, C- or N-terminus, N- termini. They can extend the chemical repertoire of the 20 standard amino acids by modifying an existing functional group or introducing a new one such as phosphorylation, phosphate. Phosphorylation is a highly effective mechanism for regulating the activity of enzymes and is the most common post-translational modification. Many eukaryotic and prokaryotic proteins also have carbohydra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endoglycosidase
An Endoglycosidase is an enzyme that releases oligosaccharides from glycoproteins or glycolipids. It may also cleave polysaccharide chains between residues that are not the terminal residue, although releasing oligosaccharides from conjugated protein and lipid molecules is more common. It breaks the glycosidic bonds between two sugar monomer in the polymer. It is different from exoglycosidase that it does not do so at the terminal residue. Hence, it is used to release long carbohydrates from conjugated molecules. If an exoglycosidase were used, every monomer in the polymer would have to be removed, one by one from the chain, taking a long time. An endoglycosidase cleaves, giving a polymeric product. PROTEIN-x1-x2-x3-x4-x5-x6-x7-x8-x9-x10-x11-...-xn Mechanism Overview The mechanism is an enzymatic hydrolysis that requires two critical molecules; a proton donor (most likely an acid) and a nucleophile(most likely a base). The Endoglycosidases mechanism has two forms; an acid catalyz ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mannose
Mannose is a sugar monomer of the aldohexose series of carbohydrates. It is a C-2 epimer of glucose. Mannose is important in human metabolism, especially in the glycosylation of certain proteins. Several congenital disorders of glycosylation are associated with mutations in enzymes involved in mannose metabolism. Mannose is not an essential nutrient; it can be produced in the human body from glucose, or converted into glucose. Mannose provides 2–5 kcal/g. It is partially excreted in the urine. Etymology The root of both "mannose" and "mannitol" is manna, which the Bible describes as the food supplied to the Israelites during their journey in the region of Sinai. Several trees and shrubs can produce a substance called manna, such as the "manna tree" ('' Fraxinus ornus'') from whose secretions mannitol was originally isolated. Structure Mannose commonly exists as two different-sized rings, the pyranose (six-membered) form and the furanose (five-membered) form. Eac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Golgi Alpha-mannosidase II
Mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase (, ''mannosidase II'', ''exo-1,3-1,6-alpha-mannosidase'', ''alpha-D-mannosidase II'', ''alpha-mannosidase II'', ''alpha1-3,6-mannosidase'', ''GlcNAc transferase I-dependent alpha1,3 lpha1,6annosidase'', ''Golgi alpha-mannosidase II'', ''ManII'', ''1,3(1,6)-alpha-D-mannosidase'', ''1,3-(1,6-)mannosyl-oligosaccharide alpha-D-mannohydrolase'') is an enzyme with systematic name ''(1->3)-(1->6)-mannosyl-oligosaccharide alpha-D-mannohydrolase''. This enzyme catalyses the following chemical reaction : Hydrolysis of the terminal (1->3)- and (1->6)-linked alpha-D-mannose residues in the mannosyl-oligosaccharide Man5(GlcNAc)3 This enzyme is involved in the synthesis of glycoproteins. It is a key enzyme of N-linked Glycan processing and is inhibited by small molecule swainsonine Swainsonine is an indolizidine alkaloid. It is a potent inhibitor of Golgi alpha-mannosidase II, an immunomodulator, and a potential chemotherapy drug. As a to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligosaccharyl Transferase
Oligosaccharyltransferase or OST () is a membrane protein complex that transfers a 14-sugar oligosaccharide from dolichol to nascent protein. It is a type of glycosyltransferase. The sugar Glc3Man9GlcNAc2 (where Glc=Glucose, Man=Mannose, and GlcNAc= ''N''-acetylglucosamine) is attached to an asparagine (Asn) residue in the sequence Asn-X-Ser or Asn-X- Thr where X is any amino acid except proline. This sequence is called a glycosylation ''sequon.'' The reaction catalyzed by OST is the central step in the ''N''-linked glycosylation pathway. Location OST is a component of the translocon in the endoplasmic reticulum (ER) membrane. A lipid-linked core-oligosaccharide is assembled at the membrane of the endoplasmic reticulum and transferred to selected asparagine residues of nascent polypeptide chains by the oligosaccharyl transferase complex. The active site of OST is located about 4 nm from the lumenal face of the ER membrane. It usually acts during translation as the nasce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
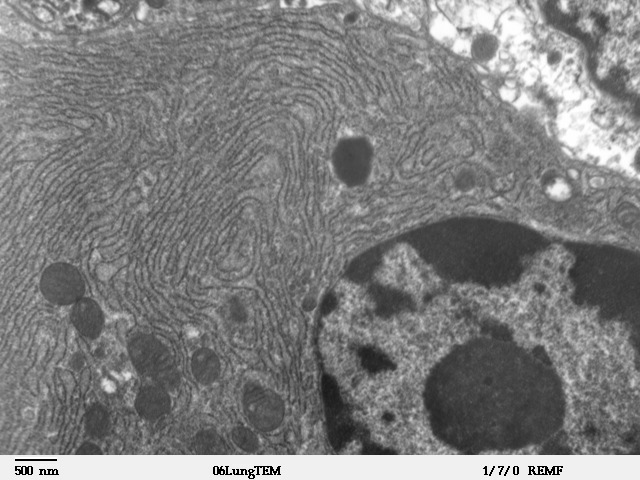
Endoplasmic Reticulum
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum (RER), and smooth endoplasmic reticulum (SER). The endoplasmic reticulum is found in most eukaryotic cells and forms an interconnected network of flattened, membrane-enclosed sacs known as cisternae (in the RER), and tubular structures in the SER. The membranes of the ER are continuous with the outer nuclear membrane. The endoplasmic reticulum is not found in red blood cells, or spermatozoa. The two types of ER share many of the same proteins and engage in certain common activities such as the synthesis of certain lipids and cholesterol. Different types of cells contain different ratios of the two types of ER depending on the activities of the cell. RER is found mainly toward the nucleus of cell and SER towards the cell membrane or plasma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomes
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins (RPs or r-proteins). The ribosomes and associated molecules are also known as the ''translational apparatus''. Overview The sequence of DNA that encodes the sequence of the amino acids in a protein is transcribed into a messenger RNA chain. Ribosomes bind to messenger RNAs and use their sequences for determining the correct sequence of amino acids to generate a given protein. Amino acids are selected and carried to the ribosome by transfer RNA (tRNA) molecules, which enter the ribosome and bind to the messenger RNA chain via an anti-cod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translation (genetics)
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription of DNA to RNA in the cell's nucleus. The entire process is called gene expression. In translation, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later folds into an active protein and performs its functions in the cell. The ribosome facilitates decoding by inducing the binding of complementary tRNA anticodon sequences to mRNA codons. The tRNAs carry specific amino acids that are chained together into a polypeptide as the mRNA passes through and is "read" by the ribosome. Translation proceeds in three phases: # Initiation: The ribosome assembles around the target mRNA. The first tRNA is attached at the start codon. # Elongation: The last tRNA validated by the small ribosomal subunit (''accommodation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
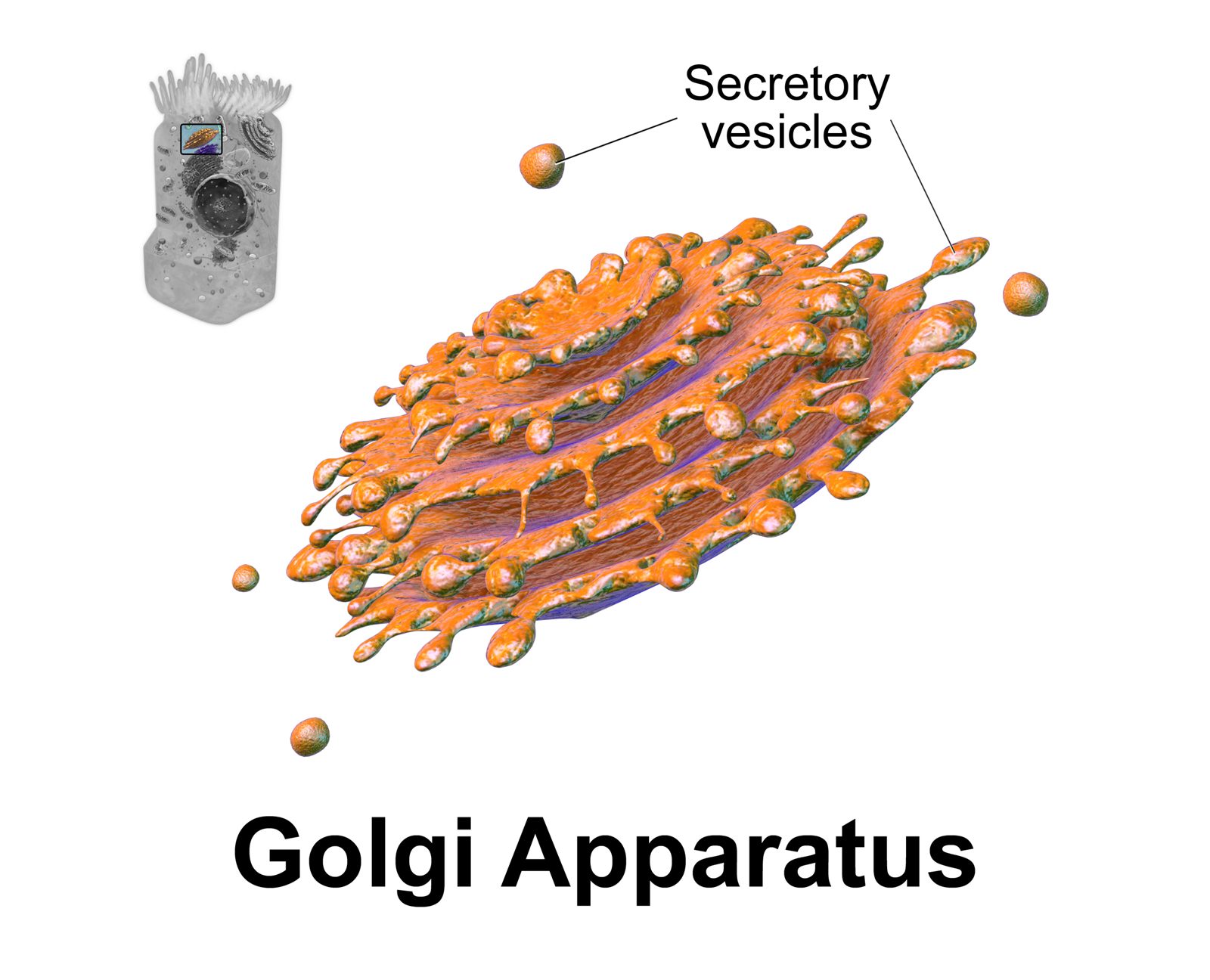
Golgi Apparatus
The Golgi apparatus (), also known as the Golgi complex, Golgi body, or simply the Golgi, is an organelle found in most eukaryotic cells. Part of the endomembrane system in the cytoplasm, it packages proteins into membrane-bound vesicles inside the cell before the vesicles are sent to their destination. It resides at the intersection of the secretory, lysosomal, and endocytic pathways. It is of particular importance in processing proteins for secretion, containing a set of glycosylation enzymes that attach various sugar monomers to proteins as the proteins move through the apparatus. It was identified in 1897 by the Italian scientist Camillo Golgi and was named after him in 1898. Discovery Owing to its large size and distinctive structure, the Golgi apparatus was one of the first organelles to be discovered and observed in detail. It was discovered in 1898 by Italian physician Camillo Golgi during an investigation of the nervous system. After first observing it under his ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-acetylglucosamine
''N''-Acetylglucosamine (GlcNAc) is an amide derivative of the monosaccharide glucose. It is a secondary amide between glucosamine and acetic acid. It is significant in several biological systems. It is part of a biopolymer in the bacterial cell wall, which is built from alternating units of GlcNAc and ''N''-acetylmuramic acid (MurNAc), cross-linked with oligopeptides at the lactic acid residue of MurNAc. This layered structure is called peptidoglycan (formerly called murein). GlcNAc is the monomeric unit of the polymer chitin, which forms the exoskeletons of arthropods like insects and crustaceans. It is the main component of the radulas of mollusks, the beaks of cephalopods, and a major component of the cell walls of most fungi. Polymerized with glucuronic acid, it forms hyaluronan. GlcNAc has been reported to be an inhibitor of elastase release from human polymorphonuclear leukocytes (range 8–17% inhibition), however this is much weaker than the inhibition seen wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
This article lists enzymes by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system. * List of EC numbers (EC 5) * List of EC numbers (EC 6) :Oxidoreductases (EC 1) (Oxidoreductase) *Dehydrogenase * Luciferase *DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) **Homoserine dehydrogenase ** Aminopropanol oxidoreductase **Diacetyl reductase **Glycerol dehydrogenase **Propanediol-phosphate dehydrogenase ** glycerol-3-phosphate dehydrogenase (NAD+) ** D-xylulose reductase **L-xylulose reductase **Lactate dehydrogenase **Malate dehydrogenase **Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) **Glucose oxidase **L-gulonolactone oxidase **Thiamine oxidase **Xanthine oxidase * :EC 1.1.4 (with a disul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |