|
Exopeptidases
An exopeptidase is any peptidase that catalyzes the cleavage of the terminal (or the penultimate) peptide bond; the process releases a single amino acid, dipeptide or a tripeptide from the peptide chain. Depending on whether the amino acid is released from the amino or the carboxy terminal (N-terminus or C-terminus), an exopeptidase is further classified as an aminopeptidase or a carboxypeptidase, respectively. Thus, an aminopeptidase, an enzyme in the brush border of the small intestine, will cleave a single amino acid from the amino terminal, whereas carboxypeptidase, which is a digestive enzyme present in pancreatic juice, will cleave a single amino acid from the carboxylic end of the peptide. Some examples of exopeptidases include: * Carboxypeptidase A - cleaves C-terminal Phe, Tyr, Trp, or Leu * Carboxypeptidase B - cleaves C-terminal Lys or Arg * Aminopeptidase - cleaves any N-terminal amino acid * Prolinase - cleaves N-terminal Pro from dipeptides * Prolidase - cleaves C-t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptidase
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Hierarchy of proteases Based on catalytic residue Proteases can be classified into seven broad groups: * Serine pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aminopeptidase
Aminopeptidases are enzymes that catalyze the cleavage of amino acids from the amino terminus (N-terminus) of proteins or peptides (exopeptidases). They are widely distributed throughout the animal and plant kingdoms and are found in many subcellular organelles, in cytosol, and as membrane components. Aminopeptidases are used in essential cellular functions. Many, but not all, of these peptidases are zinc metalloenzymes. Some aminopeptidases are monomeric, and others are assemblies of relatively high mass (50 kDa) subunits. cDNA sequences are available for several aminopeptidases and a crystal structure of the open state of human endoplasmic reticulum Aminopeptidase 1 ERAP1 is presented here. Amino acid sequences determined directly or deduced from cDNAs indicate some amino acid sequence homologies in organisms as diverse as ''Escherichia coli'' and mammals, particularly in catalytically important residues or in residues involved in metal ion binding. One important aminopeptid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Hierarchy of proteases Based on catalytic residue Proteases can be classified into seven broad groups: * Serine prot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endopeptidase
Endopeptidase or endoproteinase are proteolytic peptidases that break peptide bonds of nonterminal amino acids (i.e. within the molecule), in contrast to exopeptidases, which break peptide bonds from end-pieces of terminal amino acids. For this reason, endopeptidases cannot break down peptides into monomers, while exopeptidases can break down proteins into monomers. A particular case of endopeptidase is the oligopeptidase, whose substrates are oligopeptides instead of proteins. They are usually very specific for certain amino acids. Examples of endopeptidases include: * Trypsin - cuts after Arg or Lys, unless followed by Pro. Very strict. Works best at pH 8. * Chymotrypsin - cuts after Phe, Trp, or Tyr, unless followed by Pro. Cuts more slowly after His, Met or Leu. Works best at pH 8. * Elastase - cuts after Ala, Gly, Ser, or Val, unless followed by Pro. * Thermolysin - cuts ''before'' Ile, Met, Phe, Trp, Tyr, or Val, unless ''preceded'' by Pro. Sometimes cuts after Ala, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aminopeptidase
Aminopeptidases are enzymes that catalyze the cleavage of amino acids from the amino terminus (N-terminus) of proteins or peptides (exopeptidases). They are widely distributed throughout the animal and plant kingdoms and are found in many subcellular organelles, in cytosol, and as membrane components. Aminopeptidases are used in essential cellular functions. Many, but not all, of these peptidases are zinc metalloenzymes. Some aminopeptidases are monomeric, and others are assemblies of relatively high mass (50 kDa) subunits. cDNA sequences are available for several aminopeptidases and a crystal structure of the open state of human endoplasmic reticulum Aminopeptidase 1 ERAP1 is presented here. Amino acid sequences determined directly or deduced from cDNAs indicate some amino acid sequence homologies in organisms as diverse as ''Escherichia coli'' and mammals, particularly in catalytically important residues or in residues involved in metal ion binding. One important aminopeptid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Digestive Enzyme
Digestive enzymes are a group of enzymes that break down polymeric macromolecules into their smaller building blocks, in order to facilitate their absorption into the cells of the body. Digestive enzymes are found in the digestive tracts of animals (including humans) and in the tracts of carnivorous plants, where they aid in the digestion of food, as well as inside cells, especially in their lysosomes, where they function to maintain cellular survival. Digestive enzymes of diverse specificities are found in the saliva secreted by the salivary glands, in the secretions of cells lining the stomach, in the pancreatic juice secreted by pancreatic exocrine cells, and in the secretions of cells lining the small and large intestines. Digestive enzymes are classified based on their target substrates: *Lipases split fatty acids into fats and oils. *Proteases and peptidases split proteins into small peptides and amino acids. *Amylases split carbohydrates such as starch and sugars int ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dipeptide
A dipeptide is an organic compound derived from two amino acids. The constituent amino acids can be the same or different. When different, two isomers of the dipeptide are possible, depending on the sequence. Several dipeptides are physiologically important, and some are both physiologically and commercially significant. A well known dipeptide is aspartame, an artificial sweetener. Dipeptides are white solids. Many are far more water-soluble than the parent amino acids. For example, the dipeptide Ala-Gln has the solubility of 586 g/L more than 10x the solubility of Gln (35 g/L). Dipeptides also can exhibit different stabilities, e.g. with respect to hydrolysis. Gln does not withstand sterilization procedures, whereas this dipeptide does. Because dipeptides are prone to hydrolysis, the high solubility is exploited in infusions, i.e. to provide nutrition. Examples Commercial value About six dipeptides are of commercial interest. *Aspartame (''N''-L-α-aspartyl-L-phenyla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
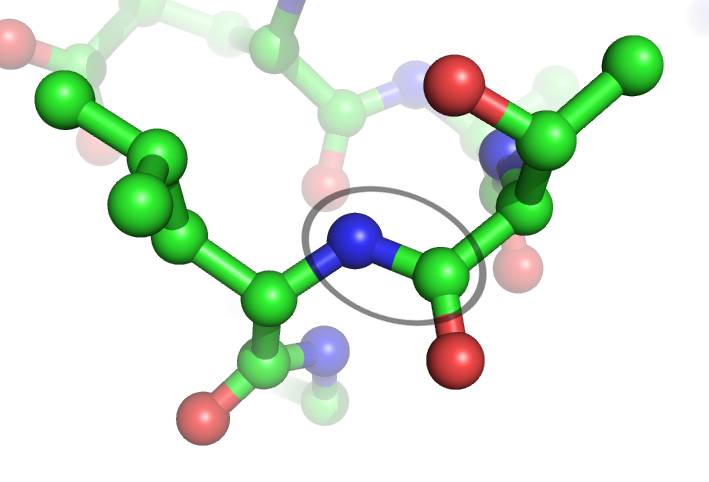
Peptide Bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 ( nitrogen number two) of another, along a peptide or protein chain. It can also be called a eupeptide bond to distinguish it from an isopeptide bond, which is another type of amide bond between two amino acids. Synthesis When two amino acids form a '' dipeptide'' through a ''peptide bond'', it is a type of condensation reaction. In this kind of condensation, two amino acids approach each other, with the non- side chain (C1) carboxylic acid moiety of one coming near the non-side chain (N2) amino moiety of the other. One loses a hydrogen and oxygen from its carboxyl group (COOH) and the other loses a hydrogen from its amino group (NH2). This reaction produces a molecule of water (H2O) and two amino acids joined by a peptide bond (−CO−NH−). The two joined amino acids are called a dipeptide. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dansyl Chloride
Dansyl chloride or 5-(DimethylAmino)Naphthalene-1-SulfonYL chloride is a reagent that reacts with primary amino groups in both aliphatic and aromatic amines to produce stable blue- or blue-green–fluorescent sulfonamide adducts. It can also be made to react with secondary amines. Dansyl chloride is widely used to modify amino acids; specifically, protein sequencing and amino acid analysis. Dansyl chloride may also be denoted DNSC. Likewise, a similar derivative, dansyl amide is known as DNSA. In addition, these protein-DNSC conjugates are sensitive to their immediate environment. This, in combination with their ability to accept energy (as in fluorescence resonance energy transfer) from the amino acid tryptophan, allows this labeling technique to be used in investigating protein folding and dynamics. The fluorescence of these sulfonamide adducts can be enhanced by adding alpha-cyclodextrin. Dansyl chloride is unstable in dimethyl sulfoxide, which should never be used to prep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Edman Degradation
Edman degradation, developed by Pehr Edman, is a method of sequencing amino acids in a peptide. In this method, the amino-terminal residue is labeled and cleaved from the peptide without disrupting the peptide bonds between other amino acid residues. Mechanism Phenyl isothiocyanate is reacted with an uncharged N-terminal amino group, under mildly alkaline conditions, to form a cyclical ''phenylthiocarbamoyl'' derivative. Then, under acidic conditions, this derivative of the terminal amino acid is cleaved as a thiazolinone derivative. The thiazolinone amino acid is then selectively extracted into an organic solvent and treated with acid to form the more stable phenylthiohydantoin (PTH)- amino acid derivative that can be identified by using chromatography or electrophoresis. This procedure can then be repeated again to identify the next amino acid. A major drawback to this technique is that the peptides being sequenced in this manner cannot have more than 50 to 60 residues ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid ''residues'' form the second-largest component ( water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling li ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |