|
Diploidization
Diploidization is the process of converting a polyploid genome back into a diploid one. Polyploidy is a product of whole genome duplication (WGD) and is followed by diploidization as a result of genome shock. The plant kingdom has undergone multiple events of polyploidization followed by diploidization in both ancient and recent lineages. It has also been hypothesized that vertebrate genomes have gone through two rounds of paleopolyploidy. The mechanisms of diploidization are poorly understood but patterns of chromosomal loss and evolution of novel genes are observed in the process. Elimination of duplicated genes Upon the formation of new polyploids, large sections of DNA are rapidly lost from one genome. The loss of DNA effectively achieves two purposes. First, the eliminated copy restores the normal gene dosage in the diploid organism. Second, the changes in chromosomal genetic structure increase the divergence of the homoeologous chromosomes (similar chromosomes from inter- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polyploid
Polyploidy is a condition in which the cells of an organism have more than one pair of ( homologous) chromosomes. Most species whose cells have nuclei ( eukaryotes) are diploid, meaning they have two sets of chromosomes, where each set contains one or more chromosomes and comes from each of two parents, resulting in pairs of homologous chromosomes between sets. However, some organisms are polyploid. Polyploidy is especially common in plants. Most eukaryotes have diploid somatic cells, but produce haploid gametes (eggs and sperm) by meiosis. A monoploid has only one set of chromosomes, and the term is usually only applied to cells or organisms that are normally diploid. Males of bees and other Hymenoptera, for example, are monoploid. Unlike animals, plants and multicellular algae have life cycles with two alternating multicellular generations. The gametophyte generation is haploid, and produces gametes by mitosis, the sporophyte generation is diploid and produces spores by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paleopolyploidy
Paleopolyploidy is the result of genome duplications which occurred at least several million years ago (MYA). Such an event could either double the genome of a single species (autopolyploidy) or combine those of two species (allopolyploidy). Because of functional redundancy, genes are rapidly silenced or lost from the duplicated genomes. Most paleopolyploids, through evolutionary time, have lost their polyploid status through a process called diploidization, and are currently considered diploids e.g. baker's yeast, ''Arabidopsis thaliana'', and perhaps humans. Paleopolyploidy is extensively studied in plant lineages. It has been found that almost all flowering plants have undergone at least one round of genome duplication at some point during their evolutionary history. Ancient genome duplications are also found in the early ancestor of vertebrates (which includes the human lineage) near the origin of the bony fishes, and another in the stem lineage of teleost fishes. Eviden ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homologous Chromosome
A couple of homologous chromosomes, or homologs, are a set of one maternal and one paternal chromosome that pair up with each other inside a cell during fertilization. Homologs have the same genes in the same loci where they provide points along each chromosome which enable a pair of chromosomes to align correctly with each other before separating during meiosis. This is the basis for Mendelian inheritance which characterizes inheritance patterns of genetic material from an organism to its offspring parent developmental cell at the given time and area. Overview Chromosomes are linear arrangements of condensed deoxyribonucleic acid (DNA) and histone proteins, which form a complex called chromatin. Homologous chromosomes are made up of chromosome pairs of approximately the same length, centromere position, and staining pattern, for genes with the same corresponding loci. One homologous chromosome is inherited from the organism's mother; the other is inherited from the organism ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
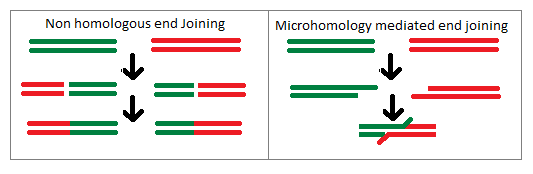
Illegitimate Recombination
Illegitimate recombination, or nonhomologous recombination, is the process by which two unrelated double stranded segments of DNA are joined. This insertion of genetic material which is not meant to be adjacent tends to lead to genes being broken causing the protein which they encode to not be properly expressed. One of the primary pathways by which this will occur is the repair mechanism known as non-homologous end joining (NHEJ). Discovery Illegitimate recombination is a natural process which was first found to be present within ''E. coli''. A 700-1400 base pair segment of DNA was found to have inserted itself into the '' gal'' and ''lac'' operons resulting in a strong polar mutation. This mechanism was then found to have the ability to insert other short genetic sequences into other locations within the bacterial genome often leading to a change in the expression of neighboring genes. Oftentimes it leads to the neighboring genes to simply shut off. However some of these segme ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neofunctionalization
Neofunctionalization, one of the possible outcomes of functional divergence, occurs when one gene copy, or paralog, takes on a totally new function after a gene duplication event. Neofunctionalization is an adaptive mutation process; meaning one of the gene copies must mutate to develop a function that was not present in the ancestral gene. In other words, one of the duplicates retains its original function, while the other accumulates molecular changes such that, in time, it can perform a different task. The process The process of Neofunctionalization begins with a gene duplication event, which is thought to occur as a defense mechanism against the accumulation of deleterious mutations. Following the gene duplication event there are two identical copies of the ancestral gene performing exactly the same function. This redundancy allows one the copies to take on a new function. In the event that the new function is advantageous, natural selection positively selects for it and the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paleopolyploidy
Paleopolyploidy is the result of genome duplications which occurred at least several million years ago (MYA). Such an event could either double the genome of a single species (autopolyploidy) or combine those of two species (allopolyploidy). Because of functional redundancy, genes are rapidly silenced or lost from the duplicated genomes. Most paleopolyploids, through evolutionary time, have lost their polyploid status through a process called diploidization, and are currently considered diploids e.g. baker's yeast, ''Arabidopsis thaliana'', and perhaps humans. Paleopolyploidy is extensively studied in plant lineages. It has been found that almost all flowering plants have undergone at least one round of genome duplication at some point during their evolutionary history. Ancient genome duplications are also found in the early ancestor of vertebrates (which includes the human lineage) near the origin of the bony fishes, and another in the stem lineage of teleost fishes. Eviden ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neofunctionalization
Neofunctionalization, one of the possible outcomes of functional divergence, occurs when one gene copy, or paralog, takes on a totally new function after a gene duplication event. Neofunctionalization is an adaptive mutation process; meaning one of the gene copies must mutate to develop a function that was not present in the ancestral gene. In other words, one of the duplicates retains its original function, while the other accumulates molecular changes such that, in time, it can perform a different task. The process The process of Neofunctionalization begins with a gene duplication event, which is thought to occur as a defense mechanism against the accumulation of deleterious mutations. Following the gene duplication event there are two identical copies of the ancestral gene performing exactly the same function. This redundancy allows one the copies to take on a new function. In the event that the new function is advantageous, natural selection positively selects for it and the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Classical Genetics
Classical genetics is the branch of genetics based solely on visible results of reproductive acts. It is the oldest discipline in the field of genetics, going back to the experiments on Mendelian inheritance by Gregor Mendel who made it possible to identify the basic mechanisms of heredity. Subsequently, these mechanisms have been studied and explained at the molecular level. Classical genetics consists of the techniques and methodologies of genetics that were in use before the advent of molecular biology. A key discovery of classical genetics in eukaryotes was genetic linkage. The observation that some genes do not segregate independently at meiosis broke the laws of Mendelian inheritance and provided science with a way to map characteristics to a location on the chromosomes. Linkage maps are still used today, especially in breeding for plant improvement. After the discovery of the genetic code and such tools of cloning as restriction enzymes, the avenues of investigation open ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |