|
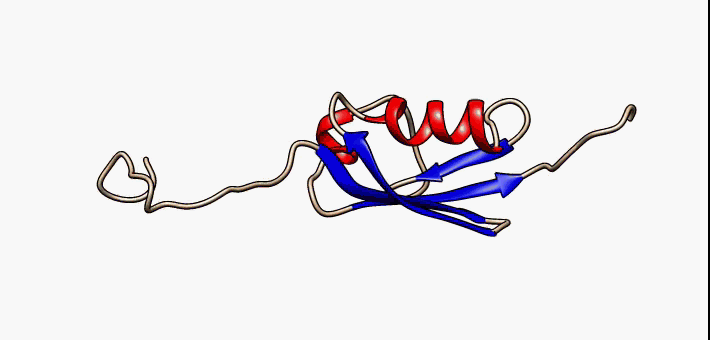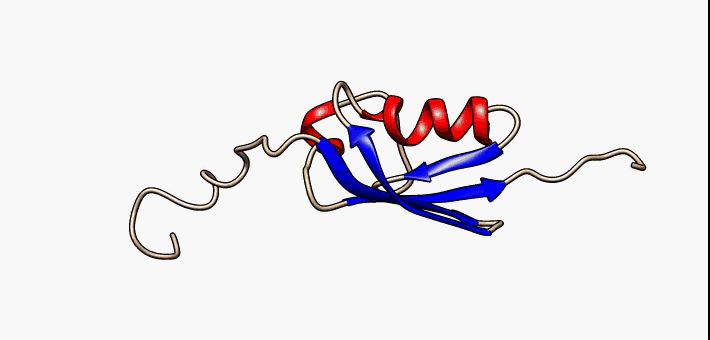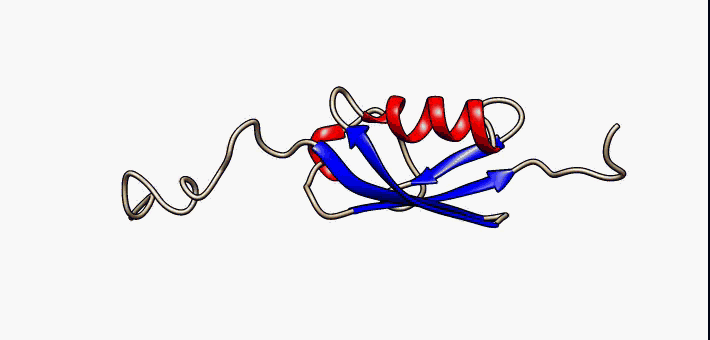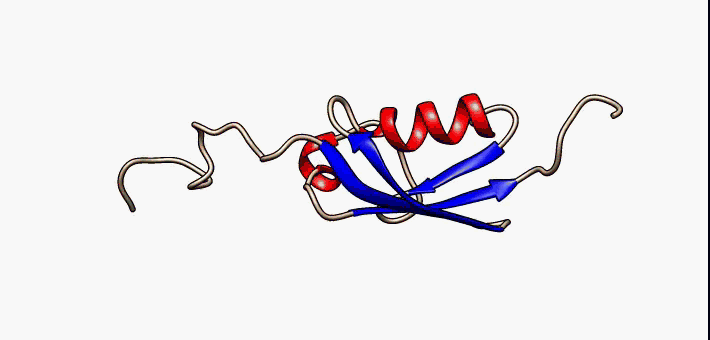
Conformational Ensemble
In computational chemistry, conformational ensembles, also known as structural ensembles, are experimentally constrained computational models describing the structure of intrinsically unstructured proteins. Such proteins are flexible in nature, lacking a stable tertiary structure, and therefore cannot be described with a single structural representation. The techniques of ensemble calculation are relatively new on the field of structural biology, and are still facing certain limitations that need to be addressed before it will become comparable to classical structural description methods such as biological macromolecular crystallography. Purpose Ensembles are models consisting of a set of conformations that together attempt to describe the structure of a flexible protein. Even though the degree of conformational freedom is extremely high, flexible/disordered protein generally differ from fully random coil structures. The main purpose of these models is to gain insights rega ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Residual Dipolar Coupling
The residual dipolar coupling between two spins in a molecule occurs if the molecules in solution exhibit a partial alignment leading to an incomplete averaging of spatially anisotropic dipolar couplings. Partial molecular alignment leads to an incomplete averaging of anisotropic magnetic interactions such as the magnetic dipole-dipole interaction (also called dipolar coupling), the chemical shift anisotropy, or the electric quadrupole interaction. The resulting so-called ''residual'' anisotropic magnetic interactions are becoming increasingly important in biomolecular NMR spectroscopy. History and pioneering works NMR spectroscopy in partially oriented media was first discovered in 1963, and in a very fundamental paper Alfred Saupe was also able to present the essential theory to describe and understand the observable phenomena only one year later. After this initiation a flood of NMR spectra in various liquid crystalline phases was reported (see ''e.g.'' ). A second technique ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intrinsically Disordered Proteins
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like aggregates, or flexible linkers in large multi-domain proteins. They are sometimes considered as a separate class of proteins along with globular, fibrous and membrane proteins. IDPs are a very large and functionally important class of proteins and their discovery has disproved the idea that three-dimensional structures of proteins must be fixed to accomplish their biological functions. For example, IDPs have been identified to participate in weak multivalent interactions that are highly cooperative and dynamic, lending them importance in DNA regulation and in cell signaling. Many IDPs can also adopt a fixed three-dimensional structure after bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Conformation
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a ''residue'' indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics. Because molecular systems typically consist of a vast number of particles, it is impossible to determine the properties of such complex systems analytically; MD simulation circumvents this problem by using numerical methods. However, long MD simulations are mathematically ill-conditioned, generating cumulative errors in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Overhauser Effect
The nuclear Overhauser effect (NOE) is the transfer of nuclear spin polarization from one population of spin-active nuclei (e.g. 1H, 13C, 15N etc.) to another via cross-relaxation. A phenomenological definition of the NOE in nuclear magnetic resonance spectroscopy (NMR) is the change in the integrated intensity (positive or negative) of one NMR resonance that occurs when another is saturated by irradiation with an RF field. The change in resonance intensity of a nucleus is a consequence of the nucleus being close in space to those directly affected by the RF perturbation. The NOE is particularly important in the assignment of NMR resonances, and the elucidation and confirmation of the structures or configurations of organic and biological molecules. The 1H two-dimensional NOE SpectroscopY (NOESY) experiment and its extensions are important tools to identify stereochemistry of proteins and other biomolecules in solution, whereas in solid form crystal x-ray diffraction typicall ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Accessible Surface Area
The accessible surface area (ASA) or solvent-accessible surface area (SASA) is the surface area of a biomolecule that is accessible to a solvent. Measurement of ASA is usually described in units of square angstroms (a standard unit of measurement in molecular biology). ASA was first described by Lee & Richards in 1971 and is sometimes called the Lee-Richards molecular surface. ASA is typically calculated using the 'rolling ball' algorithm developed by Shrake & Rupley in 1973. This algorithm uses a sphere (of solvent) of a particular radius to 'probe' the surface of the molecule. Methods of calculating ASA Shrake–Rupley algorithm The Shrake–Rupley algorithm is a numerical method that draws a mesh of points equidistant from each atom of the molecule and uses the number of these points that are solvent accessible to determine the surface area. The points are drawn at a water molecule's estimated radius beyond the van der Waals radius, which is effectively similar to ‘rolling ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogen–deuterium Exchange
Hydrogen–deuterium exchange (also called H–D or H/D exchange) is a chemical reaction in which a covalently bonded hydrogen atom is replaced by a deuterium atom, or vice versa. It can be applied most easily to exchangeable protons and deuterons, where such a transformation occurs in the presence of a suitable deuterium source, without any catalyst. The use of acid, base or metal catalysts, coupled with conditions of increased temperature and pressure, can facilitate the exchange of non-exchangeable hydrogen atoms, so long as the substrate is robust to the conditions and reagents employed. This often results in perdeuteration: hydrogen-deuterium exchange of all non-exchangeable hydrogen atoms in a molecule. An example of exchangeable protons which are commonly examined in this way are the protons of the amides in the backbone of a protein. The method gives information about the solvent accessibility of various parts of the molecule, and thus the tertiary structure of the protein. T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
J-coupling
In nuclear chemistry and nuclear physics, ''J''-couplings (also called spin-spin coupling or indirect dipole–dipole coupling) are mediated through chemical bonds connecting two spins. It is an indirect interaction between two nuclear spins that arises from hyperfine interactions between the nuclei and local electrons. In NMR spectroscopy, ''J''-coupling contains information about relative bond distances and angles. Most importantly, ''J''-coupling provides information on the connectivity of chemical bonds. It is responsible for the often complex splitting of resonance lines in the NMR spectra of fairly simple molecules. ''J''-coupling is a frequency ''difference'' that is not affected by the strength of the magnetic field, so is always stated in Hz. Vector model and manifestations for chemical structure assignments The origin of ''J''-coupling can be visualized by a vector model for a simple molecule such as hydrogen fluoride (HF). In HF, the two nuclei have spin . Four states ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Shift
In nuclear magnetic resonance (NMR) spectroscopy, the chemical shift is the resonant frequency of an atomic nucleus relative to a standard in a magnetic field. Often the position and number of chemical shifts are diagnostic of the structure of a molecule. Chemical shifts are also used to describe signals in other forms of spectroscopy such as photoemission spectroscopy. Some atomic nuclei possess a magnetic moment ( nuclear spin), which gives rise to different energy levels and resonance frequencies in a magnetic field. The total magnetic field experienced by a nucleus includes local magnetic fields induced by currents of electrons in the molecular orbitals (note that electrons have a magnetic moment themselves). The electron distribution of the same type of nucleus (e.g. ) usually varies according to the local geometry (binding partners, bond lengths, angles between bonds, and so on), and with it the local magnetic field at each nucleus. This is reflected in the spin energy le ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Computational Chemistry
Computational chemistry is a branch of chemistry that uses computer simulation to assist in solving chemical problems. It uses methods of theoretical chemistry, incorporated into computer programs, to calculate the structures and properties of molecules, groups of molecules, and solids. It is essential because, apart from relatively recent results concerning the hydrogen molecular ion (dihydrogen cation, see references therein for more details), the quantum many-body problem cannot be solved analytically, much less in closed form. While computational results normally complement the information obtained by chemical experiments, it can in some cases predict hitherto unobserved chemical phenomena. It is widely used in the design of new drugs and materials. Examples of such properties are structure (i.e., the expected positions of the constituent atoms), absolute and relative (interaction) energies, electronic charge density distributions, dipoles and higher multipole moments, vi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Small-angle X-ray Scattering
Small-angle X-ray scattering (SAXS) is a small-angle scattering technique by which nanoscale density differences in a sample can be quantified. This means that it can determine nanoparticle size distributions, resolve the size and shape of (monodisperse) macromolecules, determine pore sizes, characteristic distances of partially ordered materials, and much more. This is achieved by analyzing the elastic scattering behaviour of X-rays when travelling through the material, recording their scattering at small angles (typically 0.1 – 10°, hence the "Small-angle" in its name). It belongs to the family of small-angle scattering (SAS) techniques along with small-angle neutron scattering, and is typically done using hard X-rays with a wavelength of 0.07 – 0.2 nm.. Depending on the angular range in which a clear scattering signal can be recorded, SAXS is capable of delivering structural information of dimensions between 1 and 100 nm, and of repeat distances in partially ordered s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |