|
Chromosome Deletion
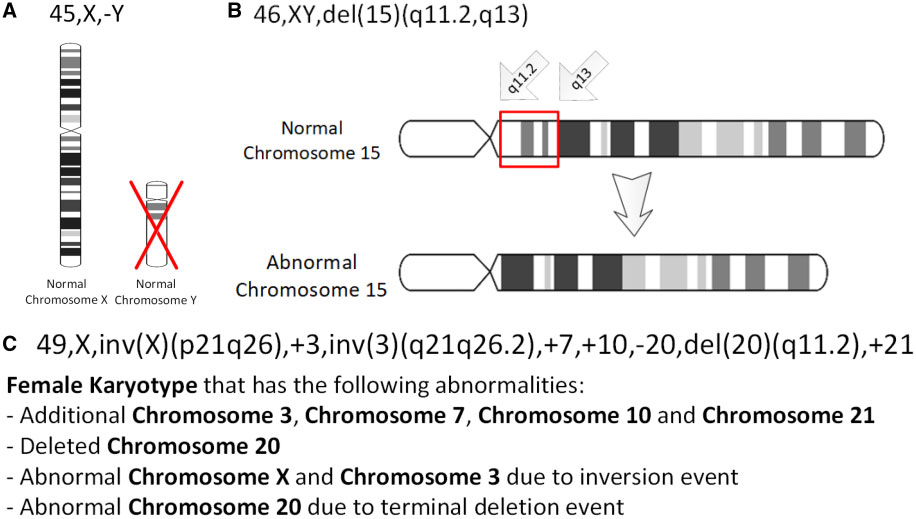
Chromosomal deletion syndromes result from deletion of parts of chromosomes. Depending on the location, size, and whom the deletion is inherited from, there are a few known different variations of chromosome deletions. Chromosomal deletion syndromes typically involve larger deletions that are visible using karyotyping techniques. Smaller deletions result in Microdeletion syndrome, which are detected using fluorescence in situ hybridization (FISH) Examples of chromosomal deletion syndromes include 5p-Deletion ( cri du chat syndrome), 4p-Deletion ( Wolf–Hirschhorn syndrome), Prader–Willi syndrome, and Angelman syndrome. 5p-Deletion The chromosomal basis of Cri du chat syndrome consists of a deletion of the most terminal portion of the short arm of chromosome 5. 5p deletions, whether terminal or interstitial, occur at different breakpoints; the chromosomal basis generally consists of a deletion on the short arm of chromosome 5. The variability seen among individuals may be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deletion (genetics)
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur which result in the deletion of a part of chromosome. The breaks can be induced by heat, viruses, radiations, chemicals. When a chromosome breaks, a part of it is deleted or lost, the missing piece of chromosome is referred to as deletion or a deficiency. For synapsis to occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into a deletion or compensation loop. The smallest single base deletion mutations occur by a single base flipping in the template DNA, followed by template DNA strand sli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Micrognathia
Micrognathism is a condition where the jaw is undersized. It is also sometimes called mandibular hypoplasia. It is common in infants, but is usually self-corrected during growth, due to the jaws' increasing in size. It may be a cause of abnormal tooth alignment and in severe cases can hamper feeding. It can also, both in adults and children, make intubation difficult, either during anesthesia or in emergency situations. Causes While not always pathological, it can present as a birth defect in multiple syndromes including: * Catel–Manzke syndrome * Bloom syndrome * Coffin–Lowry syndrome * Congenital rubella syndrome * Cri du chat syndrome * DiGeorge syndrome * Ehlers–Danlos syndrome * Fetal alcohol syndrome * Hallermann–Streiff syndrome * Hemifacial microsomia (as part of Goldenhar syndrome) * Incontinentia pigmenti * Juvenile idiopathic arthritis * Marfan syndrome * Möbius syndrome * Noonan syndrome * Pierre Robin syndrome * Prader–Willi syndrome * Progeria * Silve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Genetic Disorders
The following is a list of genetic disorders and if known, type of mutation and for the chromosome involved. Although the parlance "disease-causing gene" is common, it is the occurrence of an abnormality in the parents that causes the impairment to develop within the child. There are over 6,000 known genetic disorders in humans. Most common * P – Point mutation, or any insertion/deletion entirely inside one gene * D – Deletion of a gene or genes * Dup - Duplication of a gene or genes * C – Whole chromosome extra, missing, or both (see chromosome abnormality) * T – Trinucleotide repeat disorders: gene is extended in length Full genetic disorders list References Further reading * * * {{Medicine, state=collapsed * Disorder Genetic disorders Genetic disorders Genetic disorders A genetic disorder is a health problem caused by one or more abnormalities in the genome. It can be caused by a mutation in a single gene (monogenic) or multiple genes (polygen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hyperphagia
Polyphagia or hyperphagia is an abnormally strong, incessant sensation of hunger or desire to eat often leading to overeating. In contrast to an increase in appetite following exercise, polyphagia does not subside after eating and often leads to rapid intake of excessive quantities of food. Polyphagia is not a disorder by itself; rather, it is a symptom indicating an underlying medical condition. It is frequently a result of abnormal blood glucose levels (both hyperglycemia and hypoglycemia), and, along with polydipsia and polyuria, it is one of the "3 Ps" commonly associated with uncontrolled diabetes mellitus. Etymology and pronunciation The word ''polyphagia'' () uses combining forms of '' poly-'' + '' -phagia'', from the Greek words πολύς (polys), "very much" or "many", and φαγῶ (phago), "eating" or "devouring". Underlying conditions and possible causes Polyphagia is one of the most common symptoms of diabetes mellitus. It is associated with hyperthyroidism an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Uniparental Disomy
Uniparental disomy (UPD) occurs when a person receives two copies of a chromosome, or of part of a chromosome, from one parent and no copy from the other parent. UPD can be the result of heterodisomy, in which a pair of non-identical chromosomes are inherited from one parent (an earlier stage meiosis I error) or isodisomy, in which a single chromosome from one parent is duplicated (a later stage meiosis II error). Uniparental disomy may have clinical relevance for several reasons. For example, either isodisomy or heterodisomy can disrupt parent-specific genomic imprinting, resulting in imprinting disorders. Additionally, isodisomy leads to large blocks of homozygosity, which may lead to the uncovering of recessive genes, a similar phenomenon seen in inbred children of consanguineous partners. UPD has been found to occur in about 1 in 2,000 births. Pathophysiology UPD can occur as a random event during the formation of egg cells or sperm cells or may happen in early fetal develop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CPLX1
Complexin-1 is a protein that in humans is encoded by the ''CPLX1'' gene. Function Proteins encoded by the complexin/synaphin gene family are cytosolic proteins that function in synaptic vesicle exocytosis. These proteins bind syntaxin, part of the SNAP receptor. The protein product of this gene binds to the SNAP receptor complex and disrupts it, allowing transmitter release. Interactions CPLX1 has been shown to interact with SNAP-25 and STX1A Syntaxin-1A is a protein that in humans is encoded by the ''STX1A'' gene. Function Synaptic vesicles store neurotransmitters that are released during calcium-regulated exocytosis. The specificity of neurotransmitter release requires the local .... References External links * Further reading * * * * * * * * * * {{protein-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FGFRL1 (gene)
Fibroblast growth factor receptor-like 1 is a protein that in humans is encoded by the ''FGFRL1'' gene. The protein encoded by this gene is a member of the fibroblast growth factor receptor A fibroblast is a type of biological cell that synthesizes the extracellular matrix and collagen, produces the structural framework ( stroma) for animal tissues, and plays a critical role in wound healing. Fibroblasts are the most common cells of ... (FGFR) family, where amino acid sequence is highly conserved between members and throughout evolution. FGFR family members differ from one another in their ligand affinities and tissue distribution. A full-length representative protein would consist of an extracellular region, composed of three immunoglobulin-like domains, a single hydrophobic membrane-spanning segment and a cytoplasmic tyrosine kinase domain. The extracellular portion of the protein interacts with fibroblast growth factors, setting in motion a cascade of downstream signals, ulti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LETM1
Leucine zipper-EF-hand containing transmembrane protein 1 is a protein that in humans is encoded by the ''LETM1'' gene. Structure The LETM1 protein has a transmembrane domain and a casein kinase 2 and protein kinase C phosphorylation site. The LETM1 gene is expressed in the mitochondria of many eukaryotes indicating that this is a conserved mitochondrial protein. Function LETM1 is a eukaryotic protein that is expressed in the inner membrane of mitochondria. Experiments performed with human cells have been interpreted to indicate that it functions as a component of a Ca2+/ H+ antiporter. Experimental results with yeast cells have been interpreted as suggesting that LETM1 functions as a component of a K+/H+ antiporter. The ''Drosophila melanogaster'' LETM1 protein has been shown to functionally substitute for the K+/H+ antiporter function in yeast cells. Clinical significance Deletion of LETM1 is thought to be involved in the development of Wolf–Hirschhorn syndrome ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SLBP
Histone RNA hairpin-binding protein or stem-loop binding protein (SLBP) is a protein that in humans is encoded by the ''SLBP'' gene. Species distribution SLBP has been cloned from humans, ''C. elegans'', ''D. melanogaster'', '' X. laevis'', and sea urchins. The full length human protein has 270 amino acids (31 kDa) with a centrally located RNA binding domain (RBD). The 75 amino acid RBD is well conserved across species, however the remainder of SLBP is highly divergent in most organisms and not homologous to any other protein in the eukaryotic genomes. Function This gene encodes a protein that binds to the histone 3' UTR stem-loop structure in replication-dependent histone mRNAs. Histone mRNAs do not contain introns or polyadenylation signals, and are processed by a single endonucleolytic cleavage event downstream of the stem-loop. The stem-loop structure is essential for efficient processing of the histone pre-mRNA but this structure also controls the transport, transl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WHSC2
Negative elongation factor A is a protein that in humans is encoded by the ''WHSC2'' gene. Function This gene is expressed ubiquitously with higher levels in fetal than in adult tissues. It encodes a protein sharing 93% sequence identity with the mouse protein. Wolf-Hirschhorn syndrome (WHS) is a malformation syndrome associated with a hemizygous deletion of the distal short arm of chromosome 4. This gene is mapped to the 165 kb WHS critical region, and may play a role in the phenotype of the WHS or Pitt-Rogers-Danks syndrome. The encoded protein is found to be capable of reacting with HLA-A2-restricted and tumor-specific cytotoxic T lymphocytes, suggesting a target for use in specific immunotherapy for a large number of cancer patients. This protein has also been shown to be a member of the NELF (negative elongation factor) protein complex that participates in the regulation of RNA polymerase II transcription elongation. WHSC2 encodes the NELF-A subunit of the NELF complex. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WHSC1
Probable histone-lysine N-methyltransferase NSD2 is an enzyme that in humans is encoded by the ''NSD2'' gene. This gene encodes a protein that contains four domains present in other developmental proteins: a PWWP domain, an HMG box, a SET domain, and a PHD-type zinc finger. It is expressed ubiquitously in early development. Wolf-Hirschhorn syndrome (WHS) is a malformation syndrome associated with a hemizygous deletion of the distal short arm of chromosome 4. This gene maps to the 165 kb WHS critical region and has also been involved in the chromosomal translocation t(4;14)(p16.3;q32.3) in multiple myelomas. Alternative splicing Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be ... of this gene results in multiple transcript variants encoding different isoforms. Some tran ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
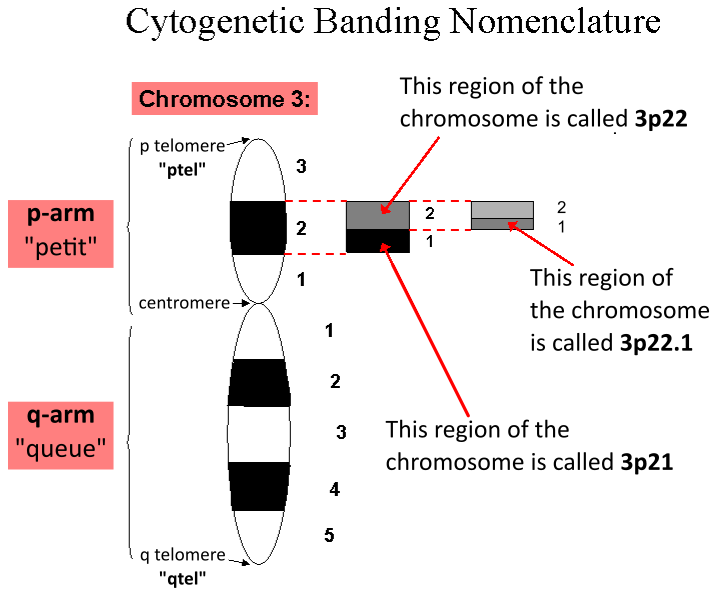
Locus (genetics)
In genetics, a locus (plural loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000. Genes may possess multiple variants known as alleles, and an allele may also be said to reside at a particular locus. Diploid and polyploid cells whose chromosomes have the same allele at a given locus are called homozygous with respect to that locus, while those that have different alleles at a given locus are called heterozygous. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the specific locus or loci responsible for producing a particular phenotype or biological trait. Association mapping, also known as "linkage disequilibrium mapping", is a method of ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |