|
Biological Age
Biomarkers of aging are biomarkers that could predict functional capacity at some later age better than chronological age. Stated another way, biomarkers of aging would give the true "biological age", which may be different from the chronological age. Validated biomarkers of aging would allow for testing interventions to extend lifespan, because changes in the biomarkers would be observable throughout the lifespan of the organism. Although maximum lifespan would be a means of validating biomarkers of aging, it would not be a practical means for long-lived species such as humans because longitudinal studies would take far too much time. Ideally, biomarkers of aging should assay the biological process of aging and not a predisposition to disease, should cause a minimal amount of trauma to assay in the organism, and should be reproducibly measurable during a short interval compared to the lifespan of the organism. An assemblage of biomarker data for an organism could be termed i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetic Mechanisms
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional genetic basis for inheritance. Epigenetics most often involves changes that affect the regulation of gene expression, but the term can also be used to describe any heritable phenotypic change. Such effects on cellular and physiological phenotypic traits may result from external or environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Gene expression c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microelectromechanical Systems
Microelectromechanical systems (MEMS), also written as micro-electro-mechanical systems (or microelectronic and microelectromechanical systems) and the related micromechatronics and microsystems constitute the technology of microscopic devices, particularly those with moving parts. They merge at the nanoscale into nanoelectromechanical systems (NEMS) and nanotechnology. MEMS are also referred to as micromachines in Japan and microsystem technology (MST) in Europe. MEMS are made up of components between 1 and 100 micrometers in size (i.e., 0.001 to 0.1 mm), and MEMS devices generally range in size from 20 micrometres to a millimetre (i.e., 0.02 to 1.0 mm), although components arranged in arrays (e.g., digital micromirror devices) can be more than 1000 mm2. They usually consist of a central unit that processes data (an integrated circuit chip such as microprocessor) and several components that interact with the surroundings (such as microsensors). Because of the la ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Senescence
Senescence () or biological aging is the gradual deterioration of functional characteristics in living organisms. The word ''senescence'' can refer to either cellular senescence or to senescence of the whole organism. Organismal senescence involves an increase in death rates and/or a decrease in fecundity with increasing age, at least in the latter part of an organism's life cycle. Senescence is the inevitable fate of almost all multicellular organisms with germ-soma separation, but it can be delayed. The discovery, in 1934, that calorie restriction can extend lifespan by 50% in rats, and the existence of species having negligible senescence and potentially immortal organisms such as '' Hydra'', have motivated research into delaying senescence and thus age-related diseases. Rare human mutations can cause accelerated aging diseases. Environmental factors may affect aging – for example, overexposure to ultraviolet radiation accelerates skin aging. Different parts of the body ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
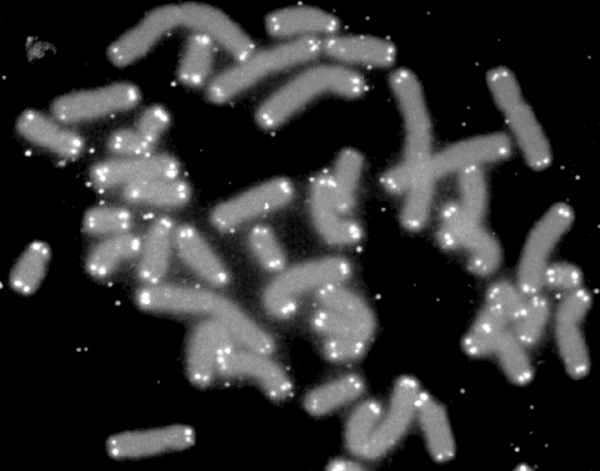
Telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes. Although there are different architectures, telomeres, in a broad sense, are a widespread genetic feature most commonly found in eukaryotes. In most, if not all species possessing them, they protect the terminal regions of chromosomal DNA from progressive degradation and ensure the integrity of linear chromosomes by preventing DNA repair systems from mistaking the very ends of the DNA strand for a double-strand break. Discovery In the early 1970s, Soviet theorist Alexei Olovnikov first recognized that chromosomes could not completely replicate their ends; this is known as the "end replication problem". Building on this, and accommodating Leonard Hayflick's idea of limited somatic cell division, Olovnikov suggested that DNA sequences are lost every time a cell replicates until the loss reaches a critical level, at which point cell division end ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cellular Senescence
Cellular senescence is a phenomenon characterized by the cessation of cell division. In their experiments during the early 1960s, Leonard Hayflick and Paul Moorhead found that normal human fetal fibroblasts in culture reach a maximum of approximately 50 cell population doublings before becoming senescent. This process is known as "replicative senescence", or the Hayflick limit. Hayflick's discovery of mortal cells paved the path for the discovery and understanding of cellular aging molecular pathways. Cellular senescence can be initiated by a wide variety of stress inducing factors. These stress factors include both environmental and internal damaging events, abnormal cellular growth, oxidative stress, autophagy factors, among many other things. The physiological importance for cell senescence has been attributed to prevention of carcinogenesis, and more recently, aging, development, and tissue repair. Senescent cells contribute to the aging phenotype, including frailty syndrom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fibroblast
A fibroblast is a type of cell (biology), biological cell that synthesizes the extracellular matrix and collagen, produces the structural framework (Stroma (tissue), stroma) for animal Tissue (biology), tissues, and plays a critical role in wound healing. Fibroblasts are the most common cells of connective tissue in animals. Structure Fibroblasts have a branched cytoplasm surrounding an elliptical, speckled cell nucleus, nucleus having two or more nucleoli. Active fibroblasts can be recognized by their abundant Endoplasmic reticulum#Rough endoplasmic reticulum, rough endoplasmic reticulum. Inactive fibroblasts (called fibrocytes) are smaller, spindle-shaped, and have a reduced amount of rough endoplasmic reticulum. Although disjointed and scattered when they have to cover a large space, fibroblasts, when crowded, often locally align in parallel clusters. Unlike the epithelial cells lining the body structures, fibroblasts do not form flat monolayers and are not restricted by a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TATA Box
In molecular biology, the TATA box (also called the Goldberg–Hogness box) is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The bacterial homolog of the TATA box is called the Pribnow box which has a shorter consensus sequence. The TATA box is considered a non-coding DNA sequence (also known as a cis-regulatory element). It was termed the "TATA box" as it contains a consensus sequence characterized by repeating T and A base pairs. How the term "box" originated is unclear. In the 1980s, while investigating nucleotide sequences in mouse genome loci, the Hogness box sequence was found and "boxed in" at the -31 position. When consensus nucleotides and alternative ones were compared, homologous regions were "boxed" by the researchers. The boxing in of sequences sheds light on the origin of the term "box". The TATA box was first identified in 1978 as a component of eukaryotic promoters. Transcription is initiated at the TATA box in TAT ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4. DNA must be compacted into nucleosomes to fit within the cell nucleus. In addition to nucleosome wrapping, eukaryotic chromatin is further compacted by being folded into a series of more complex structures, eventually forming a chromosome. Each human cell contains about 30 million nucleosomes. Nucleosomes are thought to carry epigenetically inherited information in the form of covalent modifications of their core histones. Nucleosome positions in the gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (biology)
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA ( Human genome#Coding vs. noncoding DNA), while at least 80% of mammalian genomic DNA can be actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. Transcription proceeds in the following general steps: # RNA polymerase, together with one or more general transcription factors, binds to promoter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Yeast
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are estimated to constitute 1% of all described fungal species. Yeasts are unicellular organisms that evolved from multicellular ancestors, with some species having the ability to develop multicellular characteristics by forming strings of connected budding cells known as pseudohyphae or false hyphae. Yeast sizes vary greatly, depending on species and environment, typically measuring 3–4 µm in diameter, although some yeasts can grow to 40 µm in size. Most yeasts reproduce asexually by mitosis, and many do so by the asymmetric division process known as budding. With their single-celled growth habit, yeasts can be contrasted with molds, which grow hyphae. Fungal species that can take both forms (depending on temperature or other conditions) are ca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers. There are five families of histones which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. The tight wrapping of DNA around histones ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


.jpg)

