|
Bifidobacteriales Infections
The Bifidobacteriaceae are the only family of bacteria in the order ''Bifidobacteriales''. According to the 16S rRNA-based LTP release 106 published by 'The All-Species Living Tree' Project, the order Bifidobacteriales is a clade nested within the suborder Micrococcineae, also the genus '' Bifidobacterium'' is paraphyletic to the other genera within the family, i.e. the other genera are nested within '' Bifidobacterium''. Genomics In a phylogenetic tree for the order ''Bifidobacteriales'', based on RpoB, RpoC, and DNA Gyrase B, '' Gardnerella vaginalis'' branches between different '' Bifidobacterium'' species, which makes the genus ''Bifidobacterium'' polyphyletic. The genus could be made monophyletic if ''G. vaginalis'' were placed within ''Bifidobacterium''. Comparative analysis of aligned protein sequences has led to the discovery of two conserved signature indels which are specific for the order ''Bifidobacteriales''. The first indel, a 1 amino acid deletion in ribo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bifidobacterium Adolescentis
''Bifidobacterium'' is a genus of gram-positive, nonmotile, often branched anaerobic bacteria. They are ubiquitous inhabitants of the gastrointestinal tract though strains have been isolated from the vagina and mouth ('' B. dentium'') of mammals, including humans. Bifidobacteria are one of the major genera of bacteria that make up the gastrointestinal tract microbiota in mammals. Some bifidobacteria are used as probiotics. Before the 1960s, ''Bifidobacterium'' species were collectively referred to as ''Lactobacillus bifidus''. History In 1899, Henri Tissier, a French pediatrician at the Pasteur Institute in Paris, isolated a bacterium characterised by a Y-shaped morphology ("bifid") in the intestinal microbiota of breast-fed infants and named it "bifidus". In 1907, Élie Metchnikoff, deputy director at the Pasteur Institute, propounded the theory that lactic acid bacteria are beneficial to human health. Metchnikoff observed that the longevity of Bulgarians was the result of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
16S RRNA
16S rRNA may refer to: * 16S ribosomal RNA 16 S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome ( SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure. The genes coding for it are referred to as 16S rR ..., the prokaryotic ribosomal subunit * Mitochondrially encoded 16S RNA, the eukaryotic ribosomal subunit {{Short pages monitor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Indel
Indel is a molecular biology term for an insertion or deletion of bases in the genome of an organism. It is classified among small genetic variations, measuring from 1 to 10 000 base pairs in length, including insertion and deletion events that may be separated by many years, and may not be related to each other in any way. A microindel is defined as an indel that results in a net change of 1 to 50 nucleotides. In coding regions of the genome, unless the length of an indel is a multiple of 3, it will produce a frameshift mutation. For example, a common microindel which results in a frameshift causes Bloom syndrome in the Jewish or Japanese population. Indels can be contrasted with a point mutation. An indel inserts or deletes nucleotides from a sequence, while a point mutation is a form of substitution that ''replaces'' one of the nucleotides without changing the overall number in the DNA. Indels can also be contrasted with Tandem Base Mutations (TBM), which may result from fun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conserved Signature Indels
Conserved signature inserts and deletions (CSIs) in protein sequences provide an important category of molecular markers for understanding phylogenetic relationships. CSIs, brought about by rare genetic changes, provide useful phylogenetic markers that are generally of defined size and they are flanked on both sides by conserved regions to ensure their reliability. While indels can be arbitrary inserts or deletions, CSIs are defined as only those protein indels that are present within conserved regions of the protein. The CSIs that are restricted to a particular clade or group of species, generally provide good phylogenetic markers of common evolutionary descent. Due to the rarity and highly specific nature of such changes, it is less likely that they could arise independently by either convergent or parallel evolution (i.e. homoplasy) and therefore are likely to represent synapomorphy. Other confounding factors such as differences in evolutionary rates at different sites or among ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monophyletic
In cladistics for a group of organisms, monophyly is the condition of being a clade—that is, a group of taxa composed only of a common ancestor (or more precisely an ancestral population) and all of its lineal descendants. Monophyletic groups are typically characterised by shared derived characteristics ( synapomorphies), which distinguish organisms in the clade from other organisms. An equivalent term is holophyly. The word "mono-phyly" means "one-tribe" in Greek. Monophyly is contrasted with paraphyly and polyphyly as shown in the second diagram. A ''paraphyletic group'' consists of all of the descendants of a common ancestor minus one or more monophyletic groups. A '' polyphyletic group'' is characterized by convergent features or habits of scientific interest (for example, night-active primates, fruit trees, aquatic insects). The features by which a polyphyletic group is differentiated from others are not inherited from a common ancestor. These definitions have tak ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polyphyletic
A polyphyletic group is an assemblage of organisms or other evolving elements that is of mixed evolutionary origin. The term is often applied to groups that share similar features known as homoplasies, which are explained as a result of convergent evolution. The arrangement of the members of a polyphyletic group is called a polyphyly .. ource for pronunciation./ref> It is contrasted with monophyly and paraphyly. For example, the biological characteristic of warm-bloodedness evolved separately in the ancestors of mammals and the ancestors of birds; "warm-blooded animals" is therefore a polyphyletic grouping. Other examples of polyphyletic groups are algae, C4 photosynthetic plants, and edentates. Many taxonomists aim to avoid homoplasies in grouping taxa together, with a goal to identify and eliminate groups that are found to be polyphyletic. This is often the stimulus for major revisions of the classification schemes. Researchers concerned more with ecology than with systema ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gardnerella Vaginalis
''Gardnerella vaginalis'' is a species of Gram-variable-staining facultative anaerobic bacteria. The organisms are small (1.0–1.5 μm in diameter) non-spore-forming, nonmotile coccobacilli. Once classified as ''Haemophilus vaginalis'' and afterwards as ''Corynebacterium vaginalis'', ''G. vaginalis'' grows as small, circular, convex, gray colonies on chocolate agar; it also grows on agar. A selective medium for ''G. vaginalis'' is colistin-oxolinic acid blood agar. Clinical significance ''G. vaginalis'' is a facultatively anaerobic Gram-variable rod that is involved, together with many other bacteria, mostly anaerobic, in bacterial vaginosis in some women as a result of a disruption in the normal vaginal microflora. The resident facultative anaerobic ''Lactobacillus'' population in the vagina is responsible for the acidic environment. Once the anaerobes have supplanted the normal vaginal bacteria, prescription antibiotics with anaerobic coverage may have to be given to r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
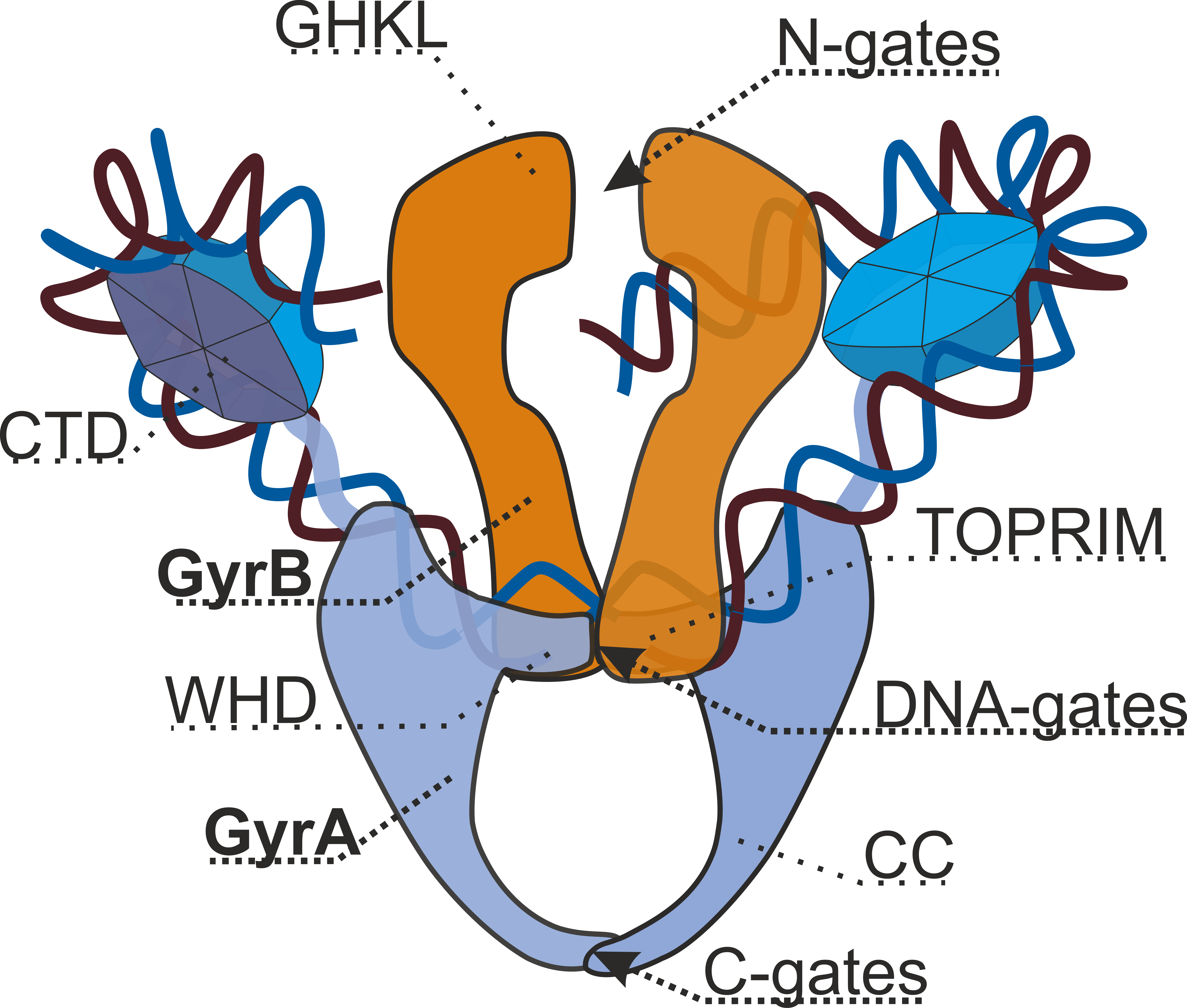
Gyrase
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. The enzyme causes negative supercoiling of the DNA or relaxes positive supercoils. It does so by looping the template so as to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite ''Plasmodium falciparum'' and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antib ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RpoB
The ''rpoB'' gene encodes the β subunit of bacterial RNA polymerase and the homologous plastid-encoded RNA polymerase (PEP). It codes for 1342 amino acids in ''E. coli'', making it the second-largest polypeptide in the bacterial cell. It is targeted by the rifamycin family of antibacterials, such as rifampin. Mutations in ''rpoB'' that confer resistance to rifamycins do so by altering the protein's drug-binding residues, thereby reducing affinity for these antibiotics. Some bacteria contain multiple copies of the 16S rRNA gene, which is commonly used as the molecular marker to study phylogeny. In these cases, the single-copy ''rpoB'' gene can be used to study microbial diversity. An inhibitor of transcription in bacteria, tagetitoxin, also inhibits PEP, showing that the complex found in plants is very similar to the homologous enzyme in bacteria. Drug resistance In a bacterium without the proper mutation(s) in ''rpoB'' rifampicin binds to a site near the fork in the β subun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic Tree
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. All life on Earth is part of a single phylogenetic tree, indicating common ancestry. In a ''rooted'' phylogenetic tree, each node with descendants represents the inferred most recent common ancestor of those descendants, and the edge lengths in some trees may be interpreted as time estimates. Each node is called a taxonomic unit. Internal nodes are generally called hypothetical taxonomic units, as they cannot be directly observed. Trees are useful in fields of biology such as bioinformatics, systematics, and phylogenetics. ''Unrooted'' trees illustrate only the relatedness of the leaf nodes and do not require the ancestral root to b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |