|
Base Calling
Base calling is the process of assigning nucleobases to chromatogram peaks, light intensity signals, or electrical current changes resulting from nucleotides passing through a nanopore. One computer program for accomplishing this job is Phred, which is a widely used base calling software program by both academic and commercial DNA sequencing laboratories because of its high base calling accuracy. Base callers for Nanopore sequencing use neural network A neural network is a network or circuit of biological neurons, or, in a modern sense, an artificial neural network, composed of artificial neurons or nodes. Thus, a neural network is either a biological neural network, made up of biological ...s trained on current signals obtained from accurate sequencing data. Base calling accuracy Base calling can be assessed by two metrics, read accuracy and consensus accuracy. Read accuracy refers to the called base's accuracy to a known reference. Consensus accuracy refers to how accu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleobase
Nucleobases, also known as ''nitrogenous bases'' or often simply ''bases'', are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic building blocks of nucleic acids. The ability of nucleobases to form base pairs and to stack one upon another leads directly to long-chain helical structures such as ribonucleic acid (RNA) and deoxyribonucleic acid (DNA). Five nucleobases— adenine (A), cytosine (C), guanine (G), thymine (T), and uracil (U)—are called ''primary'' or ''canonical''. They function as the fundamental units of the genetic code, with the bases A, G, C, and T being found in DNA while A, G, C, and U are found in RNA. Thymine and uracil are distinguished by merely the presence or absence of a methyl group on the fifth carbon (C5) of these heterocyclic six-membered rings. In addition, some viruses have aminoadenine (Z) instead of adenine. It differs in having ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatogram
In chemical analysis, chromatography is a laboratory technique for the Separation process, separation of a mixture into its components. The mixture is dissolved in a fluid solvent (gas or liquid) called the ''mobile phase'', which carries it through a system (a column, a capillary tube, a plate, or a sheet) on which a material called the ''stationary phase'' is fixed. Because the different constituents of the mixture tend to have different affinities for the stationary phase and are retained for different lengths of time depending on their interactions with its surface sites, the constituents travel at different apparent velocities in the mobile fluid, causing them to separate. The separation is based on the differential partitioning between the mobile and the stationary phases. Subtle differences in a compound's partition coefficient result in differential retention on the stationary phase and thus affect the separation. Chromatography may be preparative or analytical. The pu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nanopore
A nanopore is a pore of nanometer size. It may, for example, be created by a pore-forming protein or as a hole in synthetic materials such as silicon or graphene. When a nanopore is present in an electrically insulating membrane, it can be used as a single-molecule detector. It can be a biological protein channel in a high electrical resistance lipid bilayer, a pore in a solid-state membrane or a hybrid of these – a protein channel set in a synthetic membrane. The detection principle is based on monitoring the ionic current passing through the nanopore as a voltage is applied across the membrane. When the nanopore is of molecular dimensions, passage of molecules (e.g., DNA) cause interruptions of the "open" current level, leading to a "translocation event" signal. The passage of RNA or single-stranded DNA molecules through the membrane-embedded alpha-hemolysin channel (1.5 nm diameter), for example, causes a ~90% blockage of the current (measured at 1 M KCl solution). It m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phred (software)
Phred is a computer program for base calling, that is to say, identifying a nucleobase sequence from fluorescence "trace" data generated by an automated DNA sequencer that uses electrophoresis and 4-fluorescent dye method. When originally developed, Phred produced significantly fewer errors in the data sets examined than other methods, averaging 40–50% fewer errors. Phred quality scores have become widely accepted to characterize the quality of DNA sequences, and can be used to compare the efficacy of different sequencing methods. Background The fluorescent-dye DNA sequencing is a molecular biology technique that involves labeling single-strand DNA sequences of varied length with 4 fluorescent dyes (corresponding to 4 different bases used in DNA) and subsequently separating the DNA sequences by "slab gel"- or capillary-electrophoresis method (see DNA Sequencing). The electrophoresis run is monitored by a CCD on the DNA sequencer and this produces a time "trace" data (or "ch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neural Network
A neural network is a network or neural circuit, circuit of biological neurons, or, in a modern sense, an artificial neural network, composed of artificial neurons or nodes. Thus, a neural network is either a biological neural network, made up of biological neurons, or an artificial neural network, used for solving artificial intelligence (AI) problems. The connections of the biological neuron are modeled in artificial neural networks as weights between nodes. A positive weight reflects an excitatory connection, while negative values mean inhibitory connections. All inputs are modified by a weight and summed. This activity is referred to as a linear combination. Finally, an activation function controls the amplitude of the output. For example, an acceptable range of output is usually between 0 and 1, or it could be −1 and 1. These artificial networks may be used for predictive modeling, adaptive control and applications where they can be trained via a dataset. Self-learning re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
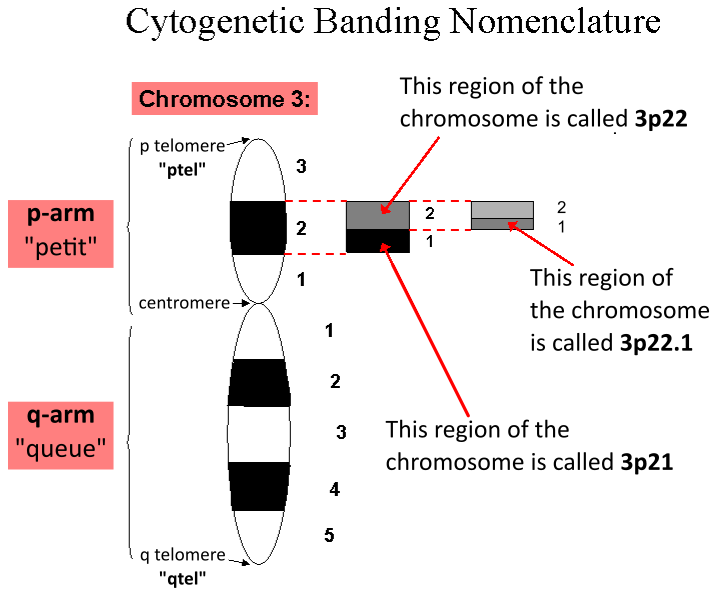
Genetic Locus
In genetics, a locus (plural loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000. Genes may possess multiple variants known as alleles, and an allele may also be said to reside at a particular locus. Diploid and polyploid cells whose chromosomes have the same allele at a given locus are called homozygous with respect to that locus, while those that have different alleles at a given locus are called heterozygous. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the specific locus or loci responsible for producing a particular phenotype or biological trait. Association mapping, also known as "linkage disequilibrium mapping", is a method of map ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


.jpg)