|
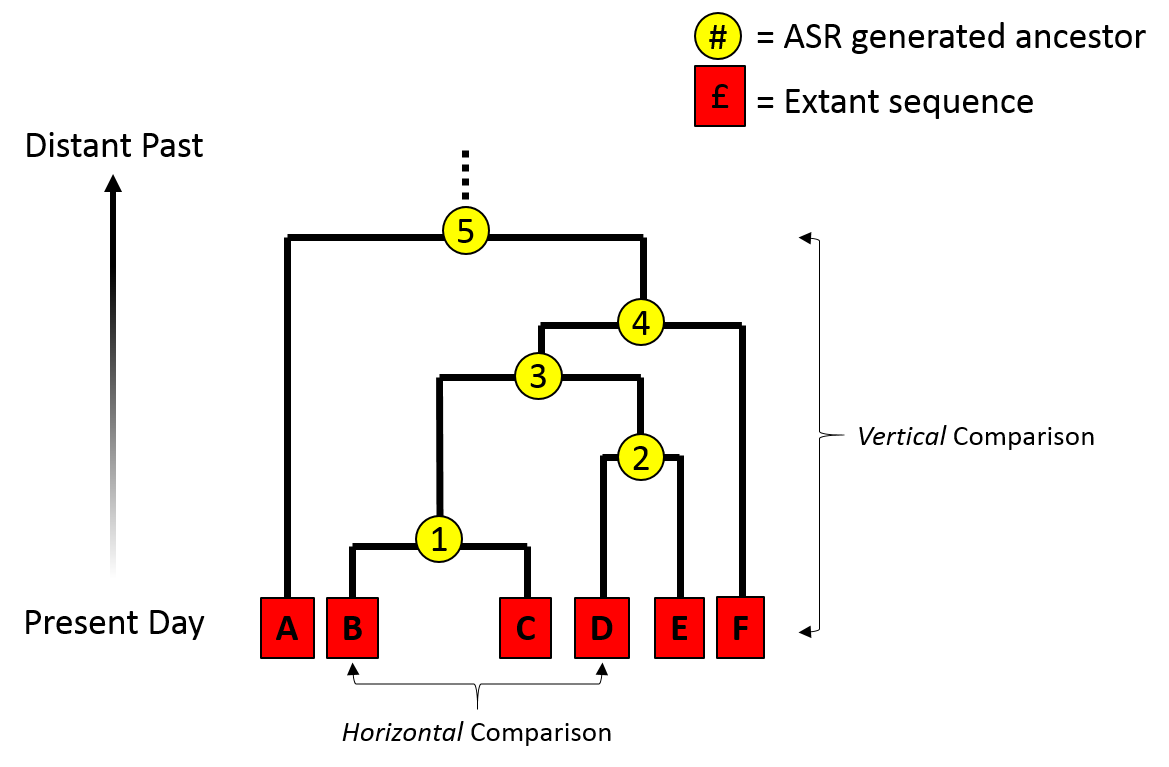
Ancestral Sequence Reconstruction
Ancestral sequence reconstruction (ASR) – also known as ancestral gene/sequence reconstruction/resurrection – is a technique used in the study of molecular evolution. The method uses related sequences to reconstruct an "ancestral" gene from a multiple sequence alignment. The method can be used to 'resurrect' ancestral proteins and was suggested in 1963 by Linus Pauling and Emile Zuckerkandl. In the case of enzymes, this approach has been called paleoenzymology (British: palaeoenzymology). Some early efforts were made in the 1980s and 1990s, led by the laboratory of Steven A. Benner, showing the potential of this technique. Thanks to the improvement of algorithms and of better sequencing and synthesis techniques, the method was developed further in the early 2000s to allow the resurrection of a greater variety of and much more ancient genes. Over the last decade, ancestral protein resurrection has developed as a strategy to reveal the mechanisms and dynamics of protein evolution ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Evolution
Molecular evolution is the process of change in the sequence composition of cellular molecules such as DNA, RNA, and proteins across generations. The field of molecular evolution uses principles of evolutionary biology and population genetics to explain patterns in these changes. Major topics in molecular evolution concern the rates and impacts of single nucleotide changes, neutral evolution vs. natural selection, origins of new genes, the genetic nature of complex traits, the genetic basis of speciation, evolution of development, and ways that evolutionary forces influence genomic and phenotypic changes. History The history of molecular evolution starts in the early 20th century with comparative biochemistry, and the use of "fingerprinting" methods such as immune assays, gel electrophoresis and paper chromatography in the 1950s to explore homologous proteins. The field of molecular evolution came into its own in the 1960s and 1970s, following the rise of molecular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic Tree
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. All life on Earth is part of a single phylogenetic tree, indicating common ancestry. In a ''rooted'' phylogenetic tree, each node with descendants represents the inferred most recent common ancestor of those descendants, and the edge lengths in some trees may be interpreted as time estimates. Each node is called a taxonomic unit. Internal nodes are generally called hypothetical taxonomic units, as they cannot be directly observed. Trees are useful in fields of biology such as bioinformatics, systematics, and phylogenetics. ''Unrooted'' trees illustrate only the relatedness of the leaf nodes and do not require the ancestral root to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymes
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
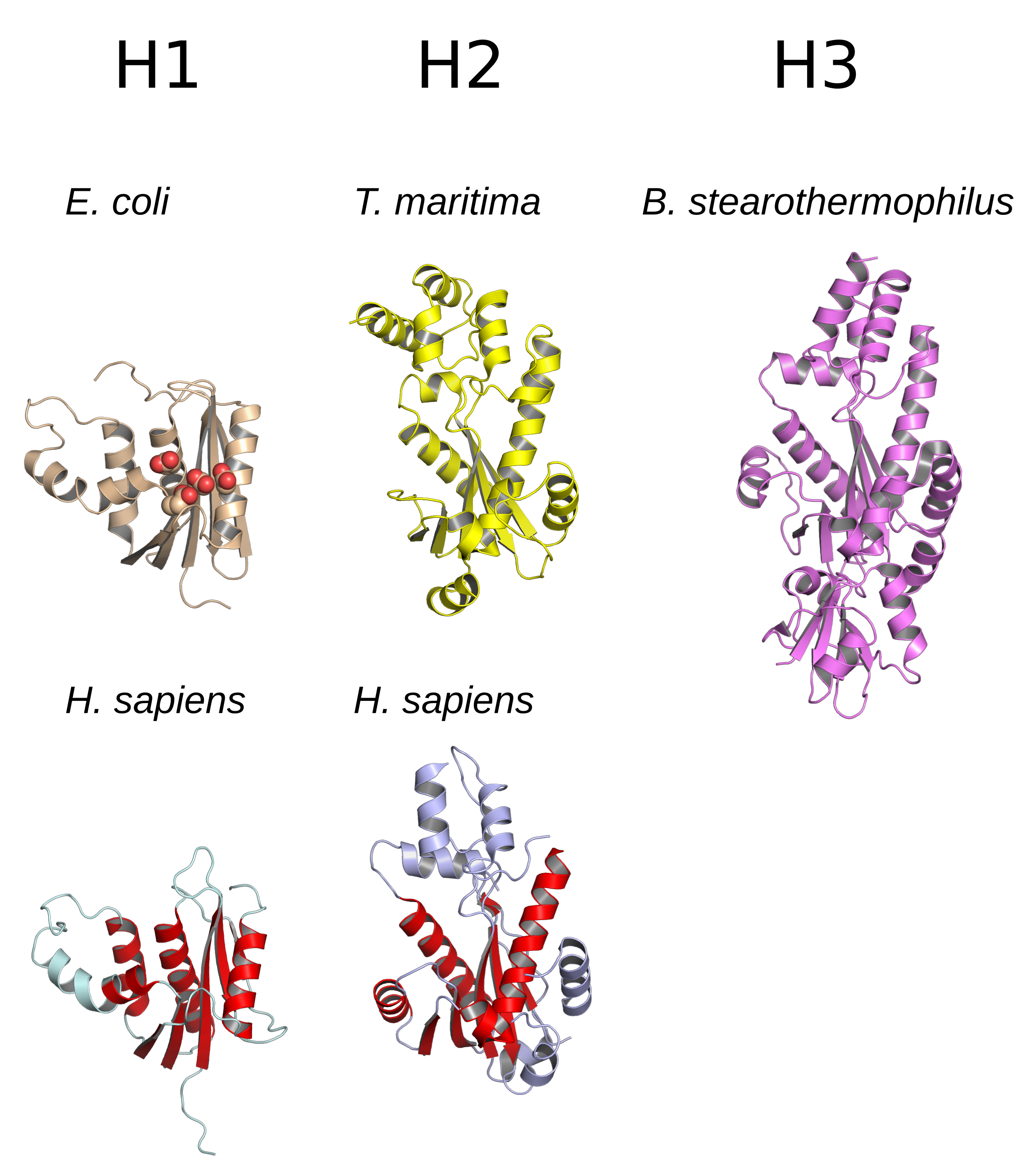
Ribonuclease H
Ribonuclease H (abbreviated RNase H or RNH) is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/ DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes. The family is divided into evolutionarily related groups with slightly different substrate preferences, broadly designated ribonuclease H1 and H2. The human genome encodes both H1 and H2. Human ribonuclease H2 is a heterotrimeric complex composed of three subunits, mutations in any of which are among the genetic causes of a rare disease known as Aicardi–Goutières syndrome. A third type, closely related to H2, is found only in a few prokaryotes, whereas H1 and H2 occur in all domains of life. Additionally, RNase H1-like retroviral ribonuclease H domains occur in multidomain reverse transcriptase proteins, which are encoded by retroviruses such as HIV and are required for vira ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Yeast
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are estimated to constitute 1% of all described fungal species. Yeasts are unicellular organisms that evolved from multicellular ancestors, with some species having the ability to develop multicellular characteristics by forming strings of connected budding cells known as pseudohyphae or false hyphae. Yeast sizes vary greatly, depending on species and environment, typically measuring 3–4 µm in diameter, although some yeasts can grow to 40 µm in size. Most yeasts reproduce asexually by mitosis, and many do so by the asymmetric division process known as budding. With their single-celled growth habit, yeasts can be contrasted with molds, which grow hyphae. Fungal species that can take both forms (depending on temperature or other conditions) a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
V-ATPase
Vacuolar-type ATPase (V-ATPase) is a highly conserved evolutionarily ancient enzyme with remarkably diverse functions in eukaryotic organisms. V-ATPases acidify a wide array of intracellular organelles and pumps protons across the plasma membranes of numerous cell types. V-ATPases couple the energy of ATP hydrolysis to proton transport across intracellular and plasma membranes of eukaryotic cells. It is generally seen as the polar opposite of ATP synthase because ATP synthase is a proton channel that uses the energy from a proton gradient to produce ATP. V-ATPase however, is a proton pump that uses the energy from ATP hydrolysis to produce a proton gradient. The Archaea-type ATPase (A-ATPase) is a related group of ATPases found in archaea that often work as an ATP synthase. It forms a clade V/A-ATPase with V-ATPase. Most members of either group shuttle protons (), but a few members have evolved to use sodium ions () instead. Roles played by V-ATPases V-ATPases are fou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hormone Receptors
A hormone receptor is a receptor molecule that binds to a specific chemical messenger . Hormone receptors are a wide family of proteins made up of receptors for thyroid and steroid hormones, retinoids and Vitamin D, and a variety of other receptors for various ligands, such as fatty acids and prostaglandins. Hormone receptors are of mainly two classes. Receptors for peptide hormones tend to be cell surface receptors built into the plasma membrane of cells and are thus referred to as trans membrane receptors. An example of this is Actrapid. Receptors for steroid hormones are usually found within the protoplasm and are referred to as intracellular or nuclear receptors, such as testosterone. Upon hormone binding, the receptor can initiate multiple signaling pathways, which ultimately leads to changes in the behavior of the target cells. Hormonal therapy and hormone receptors play a very large part in breast cancer treatment (therapy is not limited to only breast cancer). By influenci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Occam's Razor
Occam's razor, Ockham's razor, or Ocham's razor ( la, novacula Occami), also known as the principle of parsimony or the law of parsimony ( la, lex parsimoniae), is the problem-solving principle that "entities should not be multiplied beyond necessity". It is generally understood in the sense that with competing theories or explanations, the simpler one, for example a model with fewer parameters, is to be preferred. The idea is frequently attributed to English Franciscan friar William of Ockham (), a scholastic philosopher and theologian, although he never used these exact words. This philosophical razor advocates that when presented with competing hypotheses about the same prediction, one should select the solution with the fewest assumptions, and that this is not meant to be a way of choosing between hypotheses that make different predictions. Similarly, in science, Occam's razor is used as an abductive heuristic in the development of theoretical models rather than as a rigoro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Models Of DNA Evolution
A number of different Markov models of DNA sequence evolution have been proposed. These substitution models differ in terms of the parameters used to describe the rates at which one nucleotide replaces another during evolution. These models are frequently used in molecular phylogenetic analyses. In particular, they are used during the calculation of likelihood of a tree (in Bayesian and maximum likelihood approaches to tree estimation) and they are used to estimate the evolutionary distance between sequences from the observed differences between the sequences. Introduction These models are phenomenological descriptions of the evolution of DNA as a string of four discrete states. These Markov models do not explicitly depict the mechanism of mutation nor the action of natural selection. Rather they describe the relative rates of different changes. For example, mutational biases and purifying selection favoring conservative changes are probably both responsible for the relatively ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Maximum Parsimony (phylogenetics)
In phylogenetics, maximum parsimony is an optimality criterion under which the phylogenetic tree that minimizes the total number of character-state changes (or miminizes the cost of differentially weighted character-state changes) is preferred. Under the maximum-parsimony criterion, the optimal tree will minimize the amount of homoplasy (i.e., convergent evolution, parallel evolution, and evolutionary reversals). In other words, under this criterion, the shortest possible tree that explains the data is considered best. Some of the basic ideas behind maximum parsimony were presented by James S. Farris in 1970 and Walter M. Fitch in 1971. Maximum parsimony is an intuitive and simple criterion, and it is popular for this reason. However, although it is easy to ''score'' a phylogenetic tree (by counting the number of character-state changes), there is no algorithm to quickly ''generate'' the most-parsimonious tree. Instead, the most-parsimonious tree must be sought in "tree spac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BLAST
Blast or The Blast may refer to: *Explosion, a rapid increase in volume and release of energy in an extreme manner *Detonation, an exothermic front accelerating through a medium that eventually drives a shock front Film * ''Blast'' (1997 film), starring Andrew Divoff * ''Blast'' (2000 film), starring Liesel Matthews * ''Blast'' (2004 film), an action comedy film * ''Blast!'' (1972 film) or ''The Final Comedown'', an American drama * ''BLAST!'' (2008 film), a documentary about the BLAST telescope * ''A Blast'', a 2014 film directed by Syllas Tzoumerkas Magazines * ''Blast'' (magazine), a 1914–15 literary magazine of the Vorticist movement * ''Blast'' (U.S. magazine), a 1933–34 American short-story magazine * ''The Blast'' (magazine), a 1916–17 American anarchist periodical Music * Blast (American band), a hardcore punk band * Blast (Russian band), an indie band * ''Blast'' (album), by Holly Johnson, 1989 * ''The Blast'' (album), by Yuvan Shankar Raja, 1999 * "Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Position Weight Matrix
A position weight matrix (PWM), also known as a position-specific weight matrix (PSWM) or position-specific scoring matrix (PSSM), is a commonly used representation of motifs (patterns) in biological sequences. PWMs are often derived from a set of aligned sequences that are thought to be functionally related and have become an important part of many software tools for computational motif discovery. Background Creation Conversion of sequence to position probability matrix A PWM has one row for each symbol of the alphabet (4 rows for nucleotides in DNA sequences or 20 rows for amino acids in protein sequences) and one column for each position in the pattern. In the first step in constructing a PWM, a basic position frequency matrix (PFM) is created by counting the occurrences of each nucleotide at each position. From the PFM, a position probability matrix (PPM) can now be created by dividing that former nucleotide count at each position by the number of sequences, thereb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |





_JC69_Pij_Pii.jpg)