|
Affinity Label
Affinity labels are a class of enzyme inhibitors that covalently bind to their target causing its inactivation. The hallmark of an affinity label is the use of a targeting moiety to specifically and reversibly deliver a weakly reactive group to the enzyme that irreversibly binds to an amino acid residue. The targeting portion of the label often resembles the enzyme's natural substrate so that a similar mode of noncovalent binding is used prior to the covalent linkage. Their usefulness in medicine can be limited by the specificity of the first noncovalent binding step whereas indiscriminate action can be utilized for purposes such as affinity labeling - a technique for the validation of substrate-specific binding of compounds. These labels are not limited to enzymes but may also be designed to react with antibodies or ribozymes although this usage is less common. Although proteins such as hemoglobin do not have an active site, binding pockets can be exploited for their affinity and t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Inhibitor
An enzyme inhibitor is a molecule that binds to an enzyme and blocks its activity. Enzymes are proteins that speed up chemical reactions necessary for life, in which substrate molecules are converted into products. An enzyme facilitates a specific chemical reaction by binding the substrate to its active site, a specialized area on the enzyme that accelerates the most difficult step of the reaction. An enzyme inhibitor stops ("inhibits") this process, either by binding to the enzyme's active site (thus preventing the substrate itself from binding) or by binding to another site on the enzyme such that the enzyme's catalysis of the reaction is blocked. Enzyme inhibitors may bind reversibly or irreversibly. Irreversible inhibitors form a chemical bond with the enzyme such that the enzyme is inhibited until the chemical bond is broken. By contrast, reversible inhibitors bind non-covalently and may spontaneously leave the enzyme, allowing the enzyme to resume its function. Reve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Covalent Bond
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atoms, when they share electrons, is known as covalent bonding. For many molecules, the sharing of electrons allows each atom to attain the equivalent of a full valence shell, corresponding to a stable electronic configuration. In organic chemistry, covalent bonding is much more common than ionic bonding. Covalent bonding also includes many kinds of interactions, including σ-bonding, π-bonding, metal-to-metal bonding, agostic interactions, bent bonds, three-center two-electron bonds and three-center four-electron bonds. The term ''covalent bond'' dates from 1939. The prefix ''co-'' means ''jointly, associated in action, partnered to a lesser degree, '' etc.; thus a "co-valent bond", in essence, means that the atoms share " valence", such a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antibodies
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen. Each tip of the "Y" of an antibody contains a paratope (analogous to a lock) that is specific for one particular epitope (analogous to a key) on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can ''tag'' a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly (for example, by blocking a part of a virus that is essential for its invasion). To allow the immune system to recognize millions of different antigens, the antigen-binding sites at both tips of the antibody come in an equally wide variety. In contrast, the remainder of the antibody is relatively constant. It only occurs in a few vari ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalysis, catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or in vitro-evolved ribozymes are the cleavage or ligation of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3') can aminoacylate a GCCU-3' sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during Translation (biology), protein synthesis. They also participate in a variety of RNA processing reactions, including RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electrophile
In chemistry, an electrophile is a chemical species that forms bonds with nucleophiles by accepting an electron pair. Because electrophiles accept electrons, they are Lewis acids. Most electrophiles are positively charged, have an atom that carries a partial positive charge, or have an atom that does not have an octet of electrons. Electrophiles mainly interact with nucleophiles through addition and substitution reactions. Frequently seen electrophiles in organic syntheses include cations such as H+ and NO+, polarized neutral molecules such as HCl, alkyl halides, acyl halides, and carbonyl compounds, polarizable neutral molecules such as Cl2 and Br2, oxidizing agents such as organic peracids, chemical species that do not satisfy the octet rule such as carbenes and radicals, and some Lewis acids such as BH3 and DIBAL. Organic chemistry Addition of halogens These occur between alkenes and electrophiles, often halogens as in halogen addition reactions. Common reactions i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Effective Molarity
In chemistry, the effective molarity (denoted ''EM'') is defined as the ratio between the first-order rate constant of an intramolecular reaction and the second-order rate constant of the corresponding intermolecular reaction (''Kinetic Effective Molarity'') or the ratio between the equilibrium constant of an intramolecular reaction and the equilibrium constant of the corresponding intermolecular reaction (''Thermodynamic Effective Molarity''). EM_ = EM_ = EM has the dimension of concentration. High EM values always indicate greater ease of intramolecular processes over the corresponding intermolecular ones. Effective molarities can be used to get a deeper understanding of the effects of intramolecularity on reaction courses. In last decades, the frequency of use of effective molarity in scientific literature has shown a marked decrease, because this formalism is being progressively replaced by more important physical quantities. See also *Cyclic compound *Intramolecular reacti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Afatinib
Afatinib, sold under the brand name Gilotrif among others, is a medication used to treat non-small cell lung carcinoma (NSCLC). It belongs to the tyrosine kinase inhibitor family of medications. It is taken by mouth. It is mainly used to treat cases of NSCLC that harbour mutations in the epidermal growth factor receptor (EGFR) gene. It is on the World Health Organization's List of Essential Medicines. Medical uses It has received regulatory approval for use as a treatment for non-small cell lung cancer, although there is emerging evidence to support its use in other cancers such as breast cancer. Adverse effects Adverse effects by frequency include: ;Very common (>10% frequency): * Diarrhea (>90%) * Rash/dermatitis acneform * Stomatitis * Paronychia * Decreased appetite * Nose bleed * Itchiness * Dry skin ;Common (1–10% frequency): * Dehydration * Taste changes * Dry eye * Cystitis * Cheilitis * Fever * Runny/stuffy nose * Low amount of potassium in the blood * Con ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst (a catalyst is a substance that increases the rate of a chemical reaction). Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound. Cofactors can be divided into two types: inorganic ions and complex organic molecules called coenzymes. Coenzymes are mostly derived from vitamins and other organic essential nutrients in small amounts. (Note that some scientists limit the use of the term "cofactor" for inorganic substances; both types are included here.) Coenzymes are further divided into two types. The first is called a "prosthetic group", which consists of a coenzyme that is tightly (or even covalently) and permanently bound to a protein. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activity Based Proteomics
Activity-based proteomics, or activity-based protein profiling (ABPP) is a functional proteomic technology that uses chemical probes that react with mechanistically related classes of enzymes. Description The basic unit of ABPP is the probe, which typically consists of two elements: a reactive group (RG, sometimes called a "warhead") and a tag. Additionally, some probes may contain a binding group which enhances selectivity. The reactive group usually contains a specially designed electrophile that becomes covalently-linked to a nucleophilic residue in the active site of an active enzyme. An enzyme that is inhibited or post-translationally modified will not react with an activity-based probe. The tag may be either a reporter such as a fluorophore or an affinity label such as biotin or an alkyne or azide for use with the Huisgen 1,3-dipolar cycloaddition (also known as click chemistry). Advantages A major advantage of ABPP is the ability to monitor the availability of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
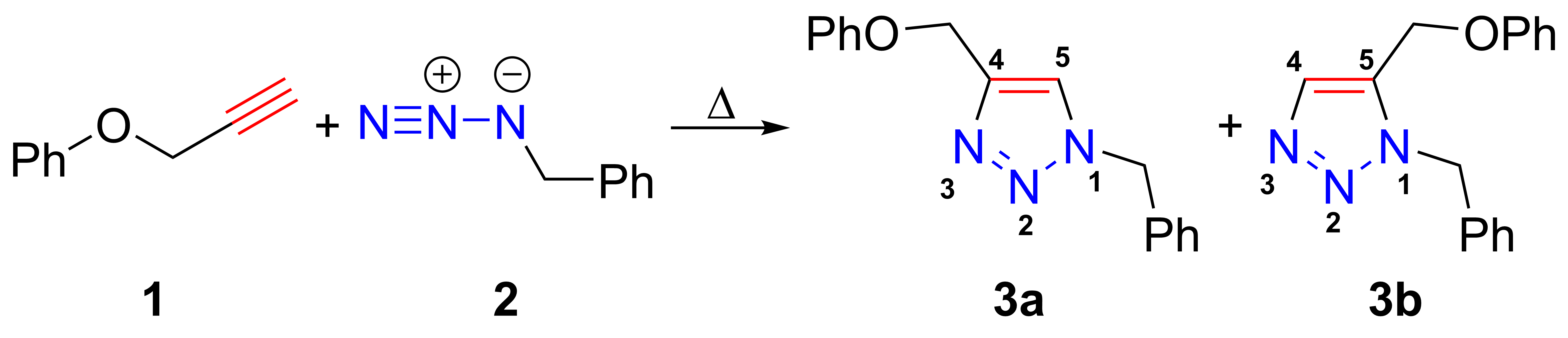
Azide-alkyne Huisgen Cycloaddition
The azide-alkyne Huisgen cycloaddition is a 1,3-dipolar cycloaddition between an azide and a terminal or internal alkyne to give a 1,2,3-triazole. Rolf Huisgen was the first to understand the scope of this organic reaction. American chemist Karl Barry Sharpless has referred to this cycloaddition as "the cream of the crop" of click chemistry and "the premier example of a click reaction". In the reaction above azide 2 reacts neatly with alkyne 1 to afford the product triazole as a mixture of 1,4-adduct (3a) and 1,5-adduct (3b) at 98 °C in 18 hours. The standard 1,3-cycloaddition between an azide 1,3-dipole and an alkene as dipolarophile has largely been ignored due to lack of reactivity as a result of electron-poor olefins and elimination side reactions. Some success has been found with non-metal-catalyzed cycloadditions, such as the reactions using dipolarophiles that are electron-poor olefins or alkynes. Although azides are not the most reactive 1,3-dipole available for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Suicide Inhibition
In biochemistry, suicide inhibition, also known as suicide inactivation or mechanism-based inhibition, is an irreversible form of enzyme inhibition that occurs when an enzyme binds a substrate analog and forms an irreversible complex with it through a covalent bond during the normal catalysis reaction. The inhibitor binds to the active site where it is modified by the enzyme to produce a reactive group that reacts irreversibly to form a stable inhibitor-enzyme complex. This usually uses a prosthetic group or a coenzyme, forming electrophilic alpha and beta unsaturated carbonyl compounds and imines. Examples Some clinical examples of suicide inhibitors include: * Disulfiram, which inhibits the acetaldehyde dehydrogenase enzyme. * Aspirin, which inhibits cyclooxygenase 1 and 2 enzymes. * Clavulanic acid, which inhibits β-lactamase: clavulanic acid covalently bonds to a serine residue in the active site of the β-lactamase, restructuring the clavulanic acid molecule, creating a muc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |