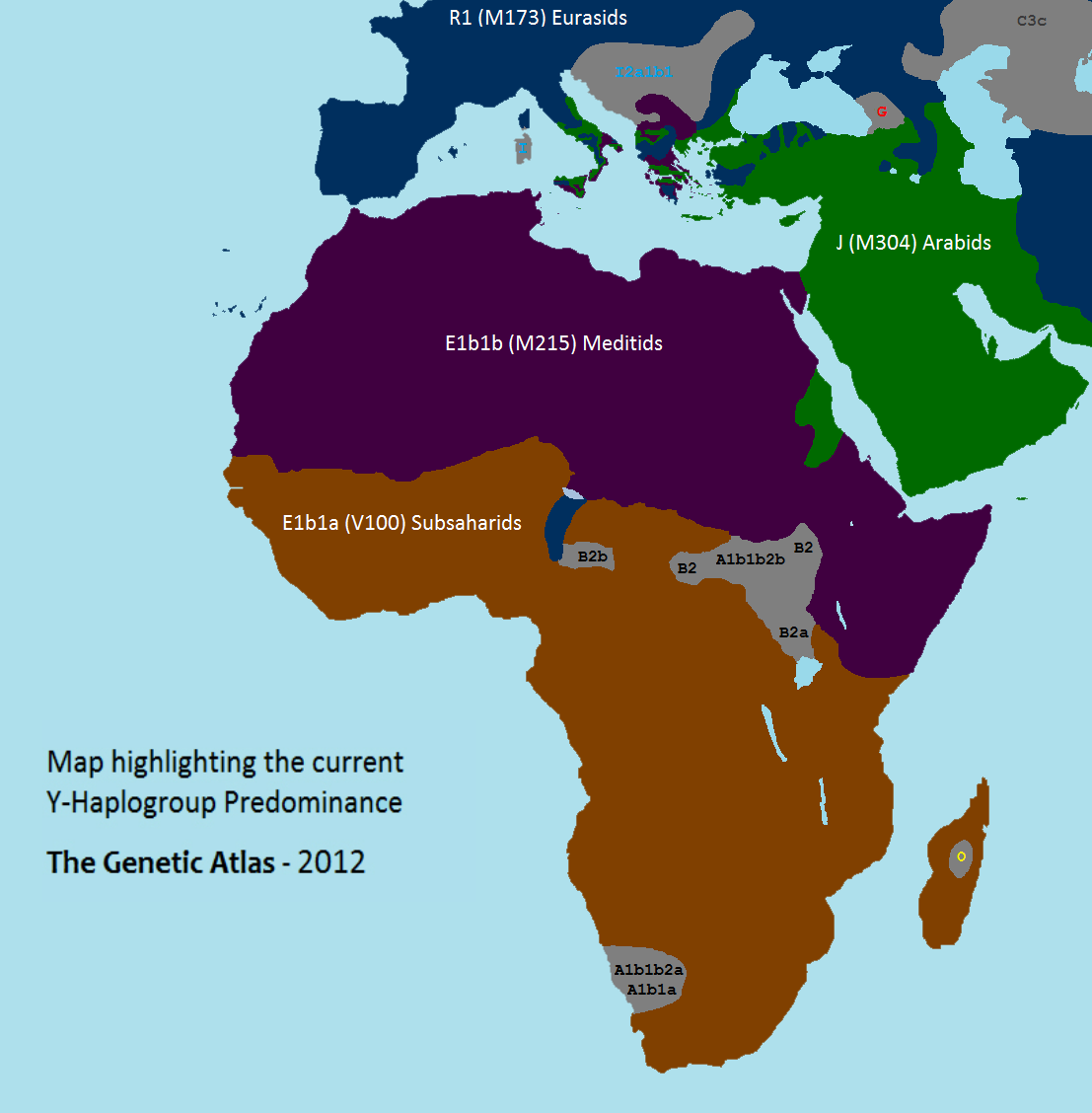
|
Y-haplogroup
In human genetics, a human Y-chromosome DNA haplogroup is a haplogroup defined by mutations in the non-genetic recombination, recombining portions of DNA from the male-specific Y chromosome (called Y-DNA). Many people within a haplogroup share similar numbers of Microsatellite, short tandem repeats (STRs) and types of mutations called single-nucleotide polymorphisms (SNPs). The human Y-chromosome accumulates roughly two mutations per generation. "one mutation in every 30 million base pairs" Y-DNA haplogroups represent major branches of the Y-chromosome phylogenetic tree that share hundreds or even thousands of mutations unique to each haplogroup. The Y-chromosomal most recent common ancestor (Y-MRCA, informally known as Y-chromosomal Adam) is the most recent common ancestor (MRCA) from whom all currently living humans are descended Patrilineality, patrilineally. Y-chromosomal Adam is estimated to have lived roughly 236,000 years ago in Africa. By examining other Population bott ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplogroup C-M130 (Y-DNA)
Haplogroup C is a major Y-chromosome haplogroup, defined by UEPs M130/RPS4Y711, P184, P255, and P260, which are all SNP mutations. It is one of two primary branches of Haplogroup CF alongside Haplogroup F. Haplogroup C is found in ancient populations on every continent except Africa and is the predominant Y-DNA haplogroup among males belonging to many peoples indigenous to East Asia, Central Asia, Siberia, North America and Australia as well as a some populations in Europe, the Levant, and later Japan.崎谷満『DNA・考古・言語の学際研究が示す新・日本列島史』(勉誠出版 2009年)(in Japanese) The haplogroup is also found with moderate to low frequency among many present-day populations of Southeast Asia, South Asia, and Southwest Asia. In addition to the basal paragroup C*, this haplogroup now has two major branches: C1 (F3393/Z1426; previously CxC3, i.e. old C1, old C2, old C4, old C5 and old C6) and C2 (M217; the former C3). Origins Haplogrou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Y-chromosomal Adam
In human genetics, the Y-chromosomal most recent common ancestor (Y-MRCA, informally known as Y-chromosomal Adam) is the patrilineal most recent common ancestor (MRCA) from whom all currently living humans are descended. He is the most recent male from whom all living humans are descended through an unbroken line of their male ancestors. The term Y-MRCA reflects the fact that the Y chromosomes of all currently living human males are directly derived from the Y chromosome of this remote ancestor. The analogous concept of the matrilineal most recent common ancestor is known as "Mitochondrial Eve" (mt-MRCA, named for the matrilineal transmission of mtDNA), the most recent woman from whom all living humans are descended matrilineally. As with "Mitochondrial Eve", the title of "Y-chromosomal Adam" is not permanently fixed to a single individual, but can advance over the course of human history as paternal lineages become extinct. Estimates of the time when Y-MRCA lived have also shifte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Marker
A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species. It can be described as a variation (which may arise due to mutation or alteration in the genomic loci) that can be observed. A genetic marker may be a short DNA sequence, such as a sequence surrounding a single base-pair change ( single nucleotide polymorphism, SNP), or a long one, like minisatellites. Background For many years, gene mapping was limited to identifying organisms by traditional phenotypes markers. This included genes that encoded easily observable characteristics such as blood types or seed shapes. The insufficient number of these types of characteristics in several organisms limited the mapping efforts that could be done. This prompted the development of gene markers which could identify genetic characteristics that are not readily observable in organisms (such as protein variation). Types Some commonly used types of genetic markers ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplogroup CF (Y-DNA)
Haplogroup CF, also known as CF-P143 and CT(xDE), is a human Y-chromosome DNA haplogroup. This paternal lineage is defined by the SNP P143. The clade's existence and distribution are inferred from the fact that haplogroups descended from CF include most human male lineages in Eurasia, Oceania and The Americas. CF is an immediate descendant of the Haplogroup CT (CT-M168), and is the sibling of Haplogroup DE (DE-YAP). It is the immediate ancestor of both Haplogroup C and Haplogroup F. There are, as yet, no confirmed cases of living individuals or human remains belonging to the basal, undivergent haplogroup CF*. In the year 2017, C-M217 (C2) & C-M130 were reported among males belonging to the Shan people The Shan people ( shn, တႆး; , my, ရှမ်းလူမျိုး; ), also known as the Tai Long, or Tai Yai are a Tai ethnic group of Southeast Asia. The Shan are the biggest minority of Burma (Myanmar) and primarily live in th ...s, who are concentrated in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplogroup E (Y-DNA)
Haplogroup E-M96 is a human Y-chromosome DNA haplogroup. It is one of the two main branches of the older and ancestral haplogroup DE, the other main branch being Haplogroup D (Y-DNA), haplogroup D. The E-M96 clade is divided into two main clades, subclades: the more common Haplogroup E-P147 (Y-DNA), E-P147, and the less common Haplogroup E-M75 (Y-DNA), E-M75. Origins Underhill (2001) proposed that haplogroup E may have arisen in East Africa. Some authors as Chandrasekar (2007), accept the earlier position of Hammer (1997) that Haplogroup E may have originated in West Asia, given that: * E is a clade of Haplogroup DE, with the other major clade, Haplogroup D (Y-DNA), haplogroup D, being exclusively distributed in Asia. * DE is a clade within Haplogroup CT (Y-DNA), M168 with the other two major clades, C and F, considered to have already a Eurasian origin. However, several discoveries made since the Hammer articles are thought to make an Asian origin less likely: # Underhill and K ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |