|
Velvet (algorithm)
Velvet is an algorithm package that has been designed to deal with ''de novo'' genome assembly and short read sequencing alignments. This is achieved through the manipulation of de Bruijn graphs for genomic sequence assembly via the removal of errors and the simplification of repeated regions.Zerbino, D. R.; Birney, E. (2008)"Velvet: de novo assembly using very short reads" Retrieved 18 October 2013. Velvet has also been implemented in commercial packages, such as Sequencher, Geneious, MacVector and BioNumerics. Introduction The development of next-generation sequencers (NGS) allowed for increased cost effectiveness on very short read sequencing. The manipulation of de Bruijn graphs as a method for alignment became more realistic but further developments were needed to address issues with errors and repeats. This led to the development of Velvet by Daniel Zerbino and Ewan Birney at the European Bioinformatics Institute in the United Kingdom. Velvet works by efficiently manipul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ewan Birney
John Frederick William Birney (known as Ewan Birney) (born 6 December 1972) is joint director of EMBL's European Bioinformatics Institute (EMBL-EBI), in Hinxton, Cambridgeshire and deputy director general of the European Molecular Biology Laboratory (EMBL). He also serves as non-executive director of Genomics England, chair of the Global Alliance for Genomics and Health (GA4GH) and honorary professor of bioinformatics at the University of Cambridge. Birney has made significant contributions to genomics, through his development of innovative bioinformatics and computational biology tools. He previously served as an associate faculty member at the Wellcome Trust Sanger Institute. Education Birney was educated at Eton College as an Oppidan Scholar. Before going to university, Birney completed a gap year internship at Cold Spring Harbor Laboratory supervised by James Watson and Adrian Krainer. Birney completed his Bachelor of Arts degree in Biochemistry at the University of Oxfor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
K-mer
In bioinformatics, ''k''-mers are substrings of length k contained within a biological sequence. Primarily used within the context of computational genomics and sequence analysis, in which ''k''-mers are composed of nucleotides (''i.e''. A, T, G, and C), ''k''-mers are capitalized upon to assemble DNA sequences, improve heterologous gene expression, identify species in metagenomic samples, and create attenuated vaccines. Usually, the term ''k''-mer refers to all of a sequence's subsequences of length k, such that the sequence AGAT would have four monomers (A, G, A, and T), three 2-mers (AG, GA, AT), two 3-mers (AGA and GAT) and one 4-mer (AGAT). More generally, a sequence of length L will have L - k + 1 ''k''-mers and n^ total possible ''k''-mers, where n is number of possible monomers (e.g. four in the case of DNA). Introduction ''k''-mers are simply length k subsequences. For example, all the possible ''k''-mers of a DNA sequence are shown below: A method of visualizi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SPAdes (software)
SPAdes (St. Petersburg genome assembler) is a genome assembly algorithm which was designed for single cell and multi-cells bacterial data sets. Therefore, it might not be suitable for large genomes projects. SPAdes works with Ion Torrent, PacBio, Oxford Nanopore, and Illumina paired-end, mate-pairs and single reads. SPAdes has been integrated into Galaxy pipelines by Guy Lionel and Philip Mabon. Background Studying the genome of single cells will help to track changes that occur in DNA over time or associated with exposure to different conditions. Additionally, many projects such as Human Microbiome Project and antibiotics discovery would greatly benefit from Single-cell sequencing (SCS). SCS has an advantage over sequencing DNA extracted from large number of cells. The problem of averaging out the significant variations between cells can be overcome by using SCS. Experimental and computational technologies are being optimized to allow researchers to sequence single cells ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
De Novo Sequence Assemblers
De novo sequence assemblers are a type of program that assembles short nucleotide sequences into longer ones without the use of a reference genome. These are most commonly used in bioinformatic studies to assemble genomes or transcriptomes. Two common types of de novo assemblers are greedy algorithm assemblers and De Bruijn graph assemblers. Types of de novo assemblers There are two types of algorithms that are commonly utilized by these assemblers: greedy, which aim for local optima, and graph method algorithms, which aim for global optima. Different assemblers are tailored for particular needs, such as the assembly of (small) bacterial genomes, (large) eukaryotic genomes, or transcriptomes. Greedy algorithm assemblers are assemblers that find local optima in alignments of smaller reads. Greedy algorithm assemblers typically feature several steps: 1) pairwise distance calculation of reads, 2) clustering of reads with greatest overlap, 3) assembly of overlapping reads into larg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with modern D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of 'junk' DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been sequenced and various regions have been annotated. The International Human Genome Project reported the sequence of the genome for ''Homo sapiens'' in 200The Human Genome Project although the initial "finished" sequence was missing 8% of the genome consisting mostly of repetitive sequences. With advancements in technology that could handle sequenci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Example 5
Example may refer to: * '' exempli gratia'' (e.g.), usually read out in English as "for example" * .example, reserved as a domain name that may not be installed as a top-level domain of the Internet ** example.com, example.net, example.org, example.edu, second-level domain names reserved for use in documentation as examples * HMS ''Example'' (P165), an Archer-class patrol and training vessel of the Royal Navy Arts * '' The Example'', a 1634 play by James Shirley * ''The Example'' (comics), a 2009 graphic novel by Tom Taylor and Colin Wilson * Example (musician) Elliot John Gleave (born 20 June 1982), better known by his stage name Example, is an English musician, singer, songwriter, rapper and record producer. His name arose due to his initials being E.G., which is an abbreviation of the Latin phrase ..., the British dance musician Elliot John Gleave (born 1982) * ''Example'' (album), a 1995 album by American rock band For Squirrels See also * * Exemplar (disambi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Example 4
Example may refer to: * '' exempli gratia'' (e.g.), usually read out in English as "for example" * .example, reserved as a domain name that may not be installed as a top-level domain of the Internet ** example.com, example.net, example.org, example.edu, second-level domain names reserved for use in documentation as examples * HMS ''Example'' (P165), an Archer-class patrol and training vessel of the Royal Navy Arts * '' The Example'', a 1634 play by James Shirley * ''The Example'' (comics), a 2009 graphic novel by Tom Taylor and Colin Wilson * Example (musician) Elliot John Gleave (born 20 June 1982), better known by his stage name Example, is an English musician, singer, songwriter, rapper and record producer. His name arose due to his initials being E.G., which is an abbreviation of the Latin phrase ..., the British dance musician Elliot John Gleave (born 1982) * ''Example'' (album), a 1995 album by American rock band For Squirrels See also * * Exemplar (disambi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
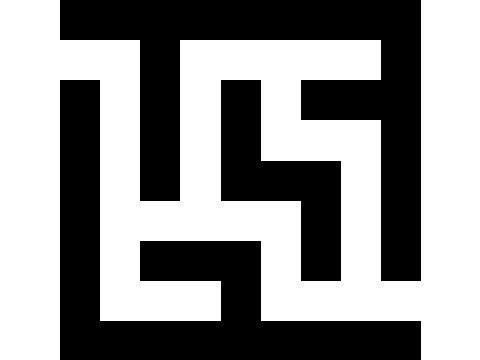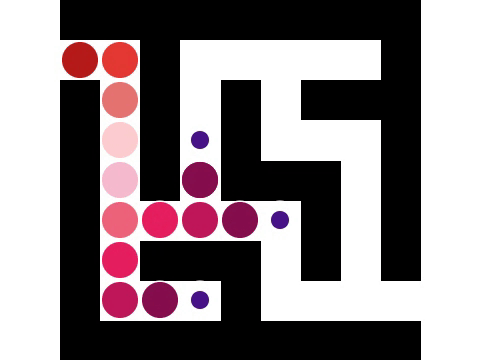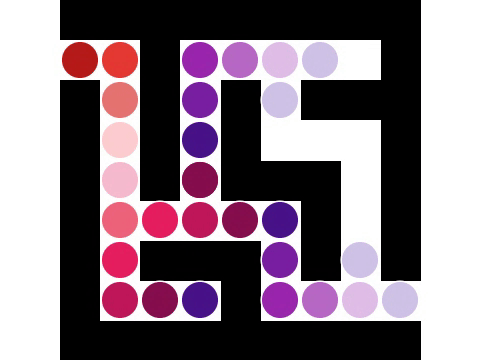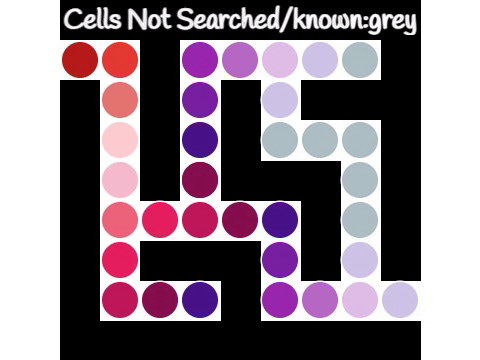
Breadth-first Search
Breadth-first search (BFS) is an algorithm for searching a tree data structure for a node that satisfies a given property. It starts at the tree root and explores all nodes at the present depth prior to moving on to the nodes at the next depth level. Extra memory, usually a queue, is needed to keep track of the child nodes that were encountered but not yet explored. For example, in a chess endgame a chess engine may build the game tree from the current position by applying all possible moves, and use breadth-first search to find a win position for white. Implicit trees (such as game trees or other problem-solving trees) may be of infinite size; breadth-first search is guaranteed to find a solution node if one exists. In contrast, (plain) depth-first search, which explores the node branch as far as possible before backtracking and expanding other nodes, may get lost in an infinite branch and never make it to the solution node. Iterative deepening depth-first search avoids ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dijkstra's Algorithm
Dijkstra's algorithm ( ) is an algorithm for finding the shortest paths between nodes in a graph, which may represent, for example, road networks. It was conceived by computer scientist Edsger W. Dijkstra in 1956 and published three years later. The algorithm exists in many variants. Dijkstra's original algorithm found the shortest path between two given nodes, but a more common variant fixes a single node as the "source" node and finds shortest paths from the source to all other nodes in the graph, producing a shortest-path tree. For a given source node in the graph, the algorithm finds the shortest path between that node and every other. It can also be used for finding the shortest paths from a single node to a single destination node by stopping the algorithm once the shortest path to the destination node has been determined. For example, if the nodes of the graph represent cities and edge path costs represent driving distances between pairs of cities connected by a dir ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Example 3
Example may refer to: * '' exempli gratia'' (e.g.), usually read out in English as "for example" * .example, reserved as a domain name that may not be installed as a top-level domain of the Internet ** example.com, example.net, example.org, example.edu, second-level domain names reserved for use in documentation as examples * HMS ''Example'' (P165), an Archer-class patrol and training vessel of the Royal Navy Arts * '' The Example'', a 1634 play by James Shirley * ''The Example'' (comics), a 2009 graphic novel by Tom Taylor and Colin Wilson * Example (musician) Elliot John Gleave (born 20 June 1982), better known by his stage name Example, is an English musician, singer, songwriter, rapper and record producer. His name arose due to his initials being E.G., which is an abbreviation of the Latin phrase ..., the British dance musician Elliot John Gleave (born 1982) * ''Example'' (album), a 1995 album by American rock band For Squirrels See also * * Exemplar (disambi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymorphism (biology)
In biology, polymorphism is the occurrence of two or more clearly different morphs or forms, also referred to as alternative ''phenotypes'', in the population of a species. To be classified as such, morphs must occupy the same habitat at the same time and belong to a panmictic population (one with random mating). Ford E.B. 1965. ''Genetic polymorphism''. Faber & Faber, London. Put simply, polymorphism is when there are two or more possibilities of a trait on a gene. For example, there is more than one possible trait in terms of a jaguar's skin colouring; they can be light morph or dark morph. Due to having more than one possible variation for this gene, it is termed 'polymorphism'. However, if the jaguar has only one possible trait for that gene, it would be termed "monomorphic". For example, if there was only one possible skin colour that a jaguar could have, it would be termed monomorphic. The term polyphenism can be used to clarify that the different forms arise from the s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |