|
Upstream Activating Sequence
An upstream activating sequence or upstream activation sequence (UAS) is a cis-acting regulatory sequence. It is distinct from the promoter and increases the expression of a neighbouring gene. Due to its essential role in activating transcription, the upstream activating sequence is often considered to be analogous to the function of the enhancer in multicellular eukaryotes. Upstream activation sequences are a crucial part of induction, enhancing the expression of the protein of interest through increased transcriptional activity. The upstream activation sequence is found adjacently upstream to a minimal promoter (TATA box) and serves as a binding site for transactivators. If the transcriptional transactivator does not bind to the UAS in the proper orientation then transcription cannot begin. To further understand the function of an upstream activation sequence, it is beneficial to see its role in the cascade of events that lead to transcription activation. The pathway begins when ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Element
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promotor (biology)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (genetics)
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA ( Human genome#Coding vs. noncoding DNA), while at least 80% of mammalian genomic DNA can be actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. Transcription proceeds in the following general steps: # RNA polymerase, together with one or more general transcription factors, binds to promoter DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enhancer (genetics)
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are ''cis''-acting. They can be located up to 1 Mbp (1,000,000 bp) away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes. The first discovery of a eukaryotic enhancer was in the immunoglobulin heavy chain gene in 1983. This enhancer, located in the large intron, provided an explanation for the transcriptional activation of rearranged Vh gene promoters while unrearranged Vh promoters remained inactive. Locations In eukaryotic cells the structure of the chromatin complex of DNA is folded in a way that functionally mimics the supercoiled state characteristic of prokaryotic DNA, so although the en ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TATA Box
In molecular biology, the TATA box (also called the Goldberg–Hogness box) is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The bacterial homolog of the TATA box is called the Pribnow box which has a shorter consensus sequence. The TATA box is considered a non-coding DNA sequence (also known as a cis-regulatory element). It was termed the "TATA box" as it contains a consensus sequence characterized by repeating T and A base pairs. How the term "box" originated is unclear. In the 1980s, while investigating nucleotide sequences in mouse genome loci, the Hogness box sequence was found and "boxed in" at the -31 position. When consensus nucleotides and alternative ones were compared, homologous regions were "boxed" by the researchers. The boxing in of sequences sheds light on the origin of the term "box". The TATA box was first identified in 1978 as a component of eukaryotic promoters. Transcription is initiated at the TATA box in TAT ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transactivator
In the context of gene regulation: transactivation is the increased rate of gene expression triggered either by biological processes or by artificial means, through the expression of an intermediate transactivator protein. In the context of receptor signaling, transactivation occurs when one or more receptors activate yet another; receptor transactivation may result from the crosstalk of signaling cascades or the activation of G protein–coupled receptor hetero-oligomer subunits, among other mechanisms. Natural transactivation Transactivation can be triggered either by endogenous cellular or viral proteins, also called transactivators. These protein factors act in trans (''i.e.'', intermolecularly). HIV and HTLV are just two of the many viruses that encode transactivators to enhance viral gene expression. These transactivators can also be linked to cancer if they start interacting with, and increasing expression of, a cellular proto-oncogene. HTLV, for instance, has been ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mediator (coactivator)
Mediator is a multiprotein complex that functions as a transcriptional coactivator in all eukaryotes. It was discovered in 1990 in the lab of Roger D. Kornberg, recipient of the 2006 Nobel Prize in Chemistry. Mediator complexes interact with transcription factors and RNA polymerase II. The main function of mediator complexes is to transmit signals from the transcription factors to the polymerase. Mediator complexes are variable at the evolutionary, compositional and conformational levels. The first image shows only one "snapshot" of what a particular mediator complex might be composed of, but it certainly does not accurately depict the conformation of the complex ''in vivo''. During evolution, mediator has become more complex. The yeast ''Saccharomyces cerevisiae'' (a simple eukaryote) is thought to have up to 21 subunits in the core mediator (exclusive of the CDK module), while mammals have up to 26. Individual subunits can be absent or replaced by other subunits under differen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase II
RNA polymerase II (RNAP II and Pol II) is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoters and begin transcription. Discovery Early studies suggested a minimum of two RNAPs: one which synthesized rRNA in the nucleolus, and one which synthesized other RNA in the nucleoplasm, part of the nucleus but outside the nucleolus. In 1969, science experimentalists Robert Roeder and William Rutter definitively discovered an additional RNAP that was responsible for transcription of some kind of RNA in the nucleoplasm. The finding was obtained by the use of ion-exchange chromatography via DEAE coated Sephadex beads. The technique separated the enzymes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
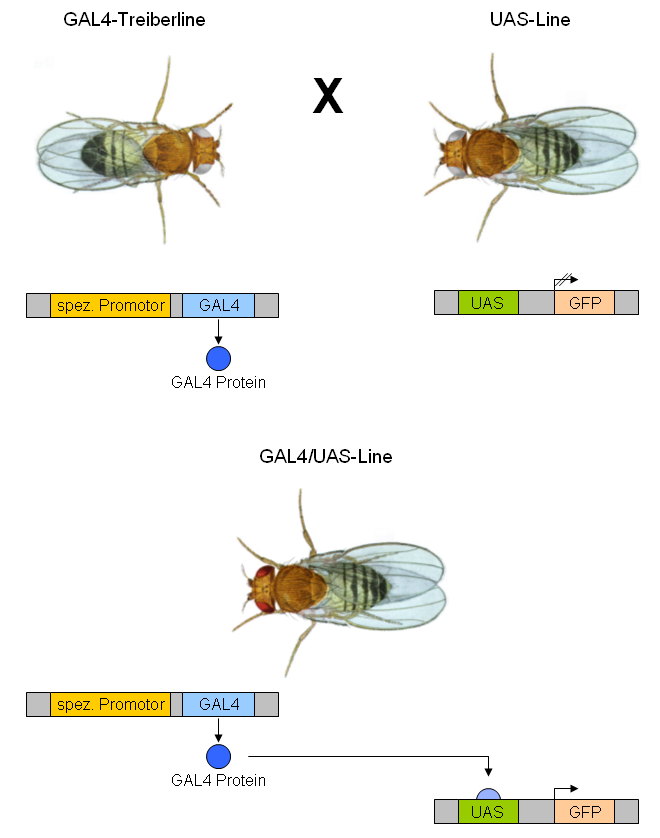
GAL4/UAS System
The GAL4-UAS system is a biochemical method used to study gene expression and function in organisms such as the fruit fly. It is based on the finding by Hitoshi Kakidani and Mark Ptashne, and Nicholas Webster and Pierre Chambon in 1988 that Gal4 binding to UAS sequences activates gene expression. The method was introduced into flies by Andrea Brand and Norbert Perrimon in 1993 and is considered a powerful technique for studying the expression of genes. The system has two parts: the Gal4 gene, encoding the yeast transcription activator protein Gal4, and the UAS (Upstream Activation Sequence), an enhancer to which GAL4 specifically binds to activate gene transcription. Overview The Gal4 system allows separation of the problems of defining which cells express a gene or protein and what the experimenter wants to do with this knowledge. Geneticists have created genetic variants of model organisms (typically fruit flies), called ''GAL4 lines'', each of which expresses GAL4 in some su ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saccharomyces Cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular biology, molecular and cell biology, much like ''Escherichia coli'' as the model bacteria, bacterium. It is the microorganism behind the most common type of fermentation (biochemistry), fermentation. ''S. cerevisiae'' cells are round to ovoid, 5–10 micrometre, μm in diameter. It reproduces by budding. Many proteins important in human biology were first discovered by studying their Homology (biology), homologs in yeast; these proteins include cell cycle proteins, signaling proteins, and protein-processing enzymes. ''S. cerevisiae'' is currently the only yeast cell known to have Berkeley body, Berkeley bo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |