|
U11 Spliceosomal RNA
The U11 snRNA ( small nuclear ribonucleic acid) is an important non-coding RNA in the minor spliceosome protein complex, which activates the alternative splicing mechanism. The minor spliceosome is associated with similar protein components as the major spliceosome. It uses U11 snRNA to recognize the 5' splice site (functionally equivalent to U1 snRNA) while U12 snRNA binds to the branchpoint to recognize the 3' splice site (functionally equivalent to U2 snRNA). Secondary structure U11 snRNA has a stem-loop structure with a 5' end as splice site sequence (5' ss) and contains four stem loops structures (I-IV). A structural comparison of U11 snRNA between plants, vertebrates and insects shows that it is folded into a structure with a four-way junction at the 5' site and in a stem loop structure at the 3' site. Binding site during assembly pathway The 5' splice site region possesses sequence complementarity with the 5' splice site of the eukaryotic U12 type pre-mRNA intr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many RNA secondary structures. As an important secondary structure of RNA, it can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reactions. Formation and stability The formation of a stem-loop structure is dependent on the stability of the resulting helix and loop regions. The first prerequisite is the presence of a sequence that can fold back on itself to form a paired double helix. The stability of this helix is determined by its length, the number of mismatches or bulges it co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tandem Repeat
Tandem repeats occur in DNA when a pattern of one or more nucleotides is repeated and the repetitions are directly adjacent to each other. Several protein domains also form tandem repeats within their amino acid primary structure, such as armadillo repeats. However, in proteins, perfect tandem repeats are unlikely in most ''in vivo'' proteins, and most known repeats are in proteins which have been designed. An example would be: : ATTCG ATTCG ATTCG in which the sequence ATTCG is repeated three times. Terminology When between 10 and 60 nucleotides are repeated, it is called a minisatellite. Those with fewer are known as microsatellites or short tandem repeats. When exactly two nucleotides are repeated, it is called a ''dinucleotide repeat'' (for example: ACACACAC...). The microsatellite instability in hereditary nonpolyposis colon cancer most commonly affects such regions. When three nucleotides are repeated, it is called a ''trinucleotide repeat'' (for example: CAGCAGCAGCAG. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
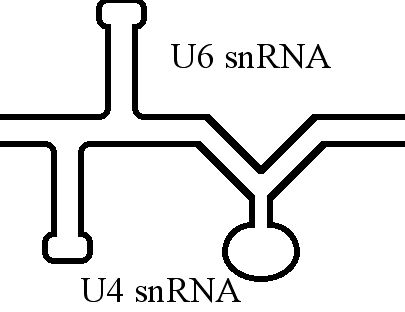
U6 Spliceosomal RNA
U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP (''small nuclear ribonucleoprotein''), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes. The RNA sequence of U6 is the most highly conserved across species of all five of the snRNAs involved in the spliceosome, suggesting that the function of the U6 snRNA has remained both crucial and unchanged through evolution. It is common in vertebrate genomes to find many copies of the U6 snRNA gene or U6-derived pseudogenes. This prevalence of "back-ups" of the U6 snRNA gene in vertebrates further implies its evolutionary importance to organism viability. The U6 snRNA gene has been isolated in many organism ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U4 Spliceosomal RNA
The U4 small nuclear Ribo-Nucleic Acid (U4 snRNA) is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6, and with each splicing round, it is displaced from the U6 snRNA (and the spliceosome) in an ATP-dependent manner, allowing U6 to re-fold and create the active site for splicing catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24 re-anneals U4 and U6. The crystal structure of a 5′ stem-loop of U4 in complex with a binding protein has been solved. Biological role The U4 snRNA has been shown to exist in a number of different formats including: bound to proteins as a small nuclear Ribo-Nuclear Protein snRNP, involved with the U6 snRNA in the di-snRNP, as well as involved with both the U6 snRNA and the U5 snRNA in the tri-snRNP. The different formats have been proposed to coincide with different temp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U6atac Minor Spliceosomal RNA
U6atac minor spliceosomal RNA is a non-coding RNA which is an essential component of the minor U12-type spliceosome complex. The U12-type spliceosome is required for removal of the rarer class of eukaryotic introns (AT-AC, U12-type). U6atac snRNA is proposed to form a base-paired complex with another spliceosomal RNA U4atac via two stem loop regions. These interacting stem loops have been shown to be required for in vivo splicing. U6atac is the functional analog of U6 spliceosomal RNA U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP (''small nuclear ribonucleoprotein''), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, ... in the major U2-type spliceosomal complex. References * External links * Non-coding RNA Spliceosome RNA splicing {{molecular-cell-biology-stub fr:ARN splicéosomal U4atac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U4atac Minor Spliceosomal RNA
U4atac minor spliceosomal RNA is a ncRNA which is an essential component of the minor U12-type spliceosome complex. The U12-type spliceosome is required for removal of the rarer class of eukaryotic introns (AT-AC, U12-type). U4atac snRNA is proposed to form a base-paired complex with another spliceosomal RNA U6atac via two stem loop regions. These interacting stem loops have been shown to be required for in vivo splicing. U4atac also contains a 3' Sm protein binding site which has been shown to be essential for splicing activity. U4atac is the functional analog of U4 spliceosomal RNA in the major U2-type spliceosomal complex. The Drosophila ''Drosophila'' () is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many speci ... U4atac snRNA has an additional predicted 3' stem loop terminal to the Sm binding si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Minor Spliceosome Mechanism
Minor may refer to: * Minor (law), a person under the age of certain legal activities. ** A person who has not reached the age of majority * Academic minor, a secondary field of study in undergraduate education Music theory *Minor chord ** Barbershop seventh chord or minor seventh chord *Minor interval *Minor key *Minor scale Mathematics * Minor (graph theory), the relation of one graph to another given certain conditions * Minor (linear algebra), the determinant of a certain submatrix People * Charles Minor (1835–1903), American college administrator * Charles A. Minor (21st-century), Liberian diplomat * Dan Minor (1909–1982), American jazz trombonist * Dave Minor (1922–1998), American basketball player * James T. Minor, US academic administrator and sociologist * Jerry Minor (born 1969), American actor, comedian and writer * Kyle Minor (born 1976), American writer * Mike Minor (actor) (born 1940), American actor * Mike Minor (baseball) (born 1987), American baseball pi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conserved Sequence
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, prompt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Precursor MRNA
Precursor or Precursors may refer to: *Precursor (religion), a forerunner, predecessor ** The Precursor, John the Baptist Science and technology * Precursor (bird), a hypothesized genus of fossil birds that was composed of fossilized parts of unrelated animals * Precursor (chemistry), a compound that participates in the chemical reaction that produces another compound * Precursor (physics), a phenomenon of wave propagation in dispersive media * Precursor in the course of a disease, a state preceding a particular stage in that course * Precursor cell (biology), a unipotent stem cell * Earthquake precursor, a diagnostic phenomenon that can occur before an earthquake * Gehrlein Precursor, a glider * LNWR Precursor Class (other), classes of passenger locomotives developed for the London and North Western Railway Fiction *Precursors Halo (series), an extremely advanced race that preceded and were destroyed by The Forerunners * ''Precursor'' (novel), a 1999 novel set in C. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U2 SnRNA
U2 spliceosomal snRNAs are a species of small nuclear RNA (snRNA) molecules found in the major spliceosomal (Sm) machinery of virtually all eukaryotic organisms. ''In vivo'', U2 snRNA along with its associated polypeptides assemble to produce the U2 small nuclear ribonucleoprotein (snRNP), an essential component of the major spliceosomal complex. The major spliceosomal-splicing pathway is occasionally referred to as U2 dependent, based on a class of Sm intron—found in mRNA primary transcripts—that are recognized exclusively by the U2 snRNP during early stages of spliceosomal assembly. In addition to U2 dependent intron recognition, U2 snRNA has been theorized to serve a catalytic role in the chemistry of pre-RNA splicing as well. Similar to ribosomal RNAs ( rRNAs), Sm snRNAs must mediate both RNA:RNA and RNA:protein contacts and hence have evolved specialized, highly conserved, primary and secondary structural elements to facilitate these types of interactions. Shortly after th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |