|
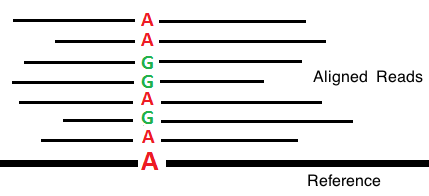
Unique Molecular Identifier
Unique molecular identifiers (UMIs), or molecular barcodes (MBC) are short sequences or molecular "tags" added to DNA fragments in some next generation sequencing library preparation protocols to identify the input DNA molecule. These tags are added before PCR amplification, and can be used to reduce errors and quantitative bias introduced by the amplification. Applications include variant calling in ctDNA, gene expression in single-cell RNA-seq (scRNA-seq) and haplotyping via linked reads. See also * Batch effect In molecular biology, a batch effect occurs when non-biological factors in an experiment cause changes in the data produced by the experiment. Such effects can lead to inaccurate conclusions when their causes are correlated with one or more outcom ... * Multiplex (assay) References DNA sequencing {{genetics-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Next Generation Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with modern DN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bias
Bias is a disproportionate weight ''in favor of'' or ''against'' an idea or thing, usually in a way that is closed-minded, prejudicial, or unfair. Biases can be innate or learned. People may develop biases for or against an individual, a group, or a belief. In science and engineering, a bias is a systematic error. Statistical bias results from an unfair sampling of a population, or from an estimation process that does not give accurate results on average. Etymology The word appears to derive from Old Provençal into Old French ''biais'', "sideways, askance, against the grain". Whence comes French ''biais'', "a slant, a slope, an oblique". It seems to have entered English via the game of bowls, where it referred to balls made with a greater weight on one side. Which expanded to the figurative use, "a one-sided tendency of the mind", and, at first especially in law, "undue propensity or prejudice". Types of bias Cognitive biases A cognitive bias is a repeating or basic m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Variant Calling
SNV calling from NGS data is any of a range of methods for identifying the existence of single nucleotide variants (SNVs) from the results of next generation sequencing (NGS) experiments. These are computational techniques, and are in contrast to special experimental methods based on known population-wide single nucleotide polymorphisms (see SNP genotyping). Due to the increasing abundance of NGS data, these techniques are becoming increasingly popular for performing SNP genotyping, with a wide variety of algorithms designed for specific experimental designs and applications. In addition to the usual application domain of SNP genotyping, these techniques have been successfully adapted to identify rare SNPs within a population, as well as detecting somatic SNVs within an individual using multiple tissue samples. Methods for detecting germline variants Most NGS based methods for SNV detection are designed to detect germline variations in the individual's genome. These are the muta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CtDNA
Circulating tumor DNA (ctDNA) is tumor-derived fragmented DNA in the bloodstream that is not associated with cells. ctDNA should not be confused with cell-free DNA (cfDNA), a broader term which describes DNA that is freely circulating in the bloodstream, but is not necessarily of tumor origin. Because ctDNA may reflect the entire tumor genome, it has gained traction for its potential clinical utility; " liquid biopsies" in the form of blood draws may be taken at various time points to monitor tumor progression throughout the treatment regimen. ctDNA originates directly from the tumor or from circulating tumor cells (CTCs), which describes viable, intact tumor cells that shed from primary tumors and enter the bloodstream or lymphatic system. The precise mechanism of ctDNA release is unclear. The biological processes postulated to be involved in ctDNA release include apoptosis and necrosis from dying cells, or active release from viable tumor cells. Studies in both human (healthy an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life— eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single Cell Sequencing
Single-cell sequencing examines the sequence information from individual cells with optimized next-generation sequencing technologies, providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment. For example, in cancer, sequencing the DNA of individual cells can give information about mutations carried by small populations of cells. In development, sequencing the RNAs expressed by individual cells can give insight into the existence and behavior of different cell types. In microbial systems, a population of the same species can appear genetically clonal. Still, single-cell sequencing of RNA or epigenetic modifications can reveal cell-to-cell variability that may help populations rapidly adapt to survive in changing environments. Background A typical human cell consists of about 2 x 3.3 billion base pairs of DNA and 600 million mRNA bases. Usually, a mix of millions of cells is used ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplotyping
A haplotype ( haploid genotype) is a group of alleles in an organism that are inherited together from a single parent. Many organisms contain genetic material ( DNA) which is inherited from two parents. Normally these organisms have their DNA organized in two sets of pairwise similar chromosomes. The offspring gets one chromosome in each pair from each parent. A set of pairs of chromosomes is called diploid and a set of only one half of each pair is called haploid. The haploid genotype (haplotype) is a genotype that considers the singular chromosomes rather than the pairs of chromosomes. It can be all the chromosomes from one of the parents or a minor part of a chromosome, for example a sequence of 9000 base pairs. However, there are other uses of this term. First, it is used to mean a collection of specific alleles (that is, specific DNA sequences) in a cluster of tightly linked genes on a chromosome that are likely to be inherited together—that is, they are likely to be co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Batch Effect
In molecular biology, a batch effect occurs when non-biological factors in an experiment cause changes in the data produced by the experiment. Such effects can lead to inaccurate conclusions when their causes are correlated with one or more outcomes of interest in an experiment. They are common in many types of high-throughput sequencing experiments, including those using microarrays, mass spectrometers, and single-cell RNA-sequencing data. They are most commonly discussed in the context of genomics and high-throughput sequencing research, but they exist in other fields of science as well. Definitions Multiple definitions of the term "batch effect" have been proposed in the literature. Lazar et al. (2013) noted, "Providing a complete and unambiguous definition of the so-called batch effect is a challenging task, especially because its origins and the way it manifests in the data are not completely known or not recorded." Focusing on microarray experiments, they propose a new def ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Multiplex (assay)
In the biological sciences, a multiplex assay is a type of immunoassay that uses magnetic beads to simultaneously measure multiple analytes in a single experiment. A multiplex assay is a derivative of an ELISA using beads for binding the capture antibody. Multiplex assays are still more common in research than in clinical settings. In a multiplex assay, microspheres of designated colors are coated with antibodies of defined binding specificities. The results can be read by flow cytometry Flow cytometry (FC) is a technique used to detect and measure physical and chemical characteristics of a population of cells or particles. In this process, a sample containing cells or particles is suspended in a fluid and injected into the fl ... because the beads are distinguishable by fluorescent signature. The number of analytes measured is determined by the number of different bead colors. Multiplex assays within a given application area or class of technology can be further stratified ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |