|
Three-finger Toxin
Three-finger toxins (abbreviated 3FTx) are a protein superfamily of small toxin proteins found in the venom of snakes. Three-finger toxins are in turn members of a larger superfamily of three-finger protein domains which includes non-toxic proteins that share a similar protein fold. The group is named for its common structure consisting of three beta strand loops connected to a central core containing four conserved disulfide bonds. The 3FP protein domain has no enzymatic activity and is typically between 60-74 amino acid residues long. Despite their conserved structure, three-finger toxin proteins have a wide range of pharmacological effects. Most members of the family are neurotoxins that act on cholinergic intercellular signaling; the alpha-neurotoxin family interacts with muscle nicotinic acetylcholine receptors (nAChRs), the kappa-bungarotoxin family with neuronal nAChRs, and muscarinic toxins with muscarinic acetylcholine receptors (mAChRs). Structure The three-finger t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Muscle-type Nicotinic Receptor
The muscle-type nicotinic receptor is a type of nicotinic acetylcholine receptor consisting of the subunit combination (α1)2β1δε (adult receptor) or (α1)2β1δγ (fetal receptor). These receptors are found in neuromuscular junctions, where activation leads to an excitatory postsynaptic potential (EPSP), mainly by increased Na+ and K+ permeability. Activation Tetraethylammonium (TEA) is a molecule found to be a weak agonist of the muscle‐type nicotinic receptor. Since receptor activation occurs as isolated bursts, it has been proposed that the receptors have a very low channel‐opening rate constant when bound to TEA. Inhibition Lidocaine, a local anesthetic, has multiple inhibitory actions on the receptor and analysis of the structure of lidocaine has identified the presence of a hydrophobic aromatic ring and a hydrophilic terminal amine. Diethylamine (DEA), a molecule that mimics the hydrophilic moiety of lidocaine by way of a positively charged amine, has been fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", '' di-'' + '' -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins. A protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein-coupled cannabinoid receptors have the ability to form both homo- and heterodimers with several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Covalent Bond
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atoms, when they share electrons, is known as covalent bonding. For many molecules, the sharing of electrons allows each atom to attain the equivalent of a full valence shell, corresponding to a stable electronic configuration. In organic chemistry, covalent bonding is much more common than ionic bonding. Covalent bonding also includes many kinds of interactions, including σ-bonding, π-bonding, metal-to-metal bonding, agostic interactions, bent bonds, three-center two-electron bonds and three-center four-electron bonds. The term ''covalent bond'' dates from 1939. The prefix ''co-'' means ''jointly, associated in action, partnered to a lesser degree, '' etc.; thus a "co-valent bond", in essence, means that the atoms share " valence", such a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monomer
In chemistry, a monomer ( ; ''mono-'', "one" + '' -mer'', "part") is a molecule that can react together with other monomer molecules to form a larger polymer chain or three-dimensional network in a process called polymerization. Classification Monomers can be classified in many ways. They can be subdivided into two broad classes, depending on the kind of the polymer that they form. Monomers that participate in condensation polymerization have a different stoichiometry than monomers that participate in addition polymerization: : Other classifications include: *natural vs synthetic monomers, e.g. glycine vs caprolactam, respectively *polar vs nonpolar monomers, e.g. vinyl acetate vs ethylene, respectively *cyclic vs linear, e.g. ethylene oxide vs ethylene glycol, respectively The polymerization of one kind of monomer gives a homopolymer. Many polymers are copolymers, meaning that they are derived from two different monomers. In the case of condensation polymerizations, the r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
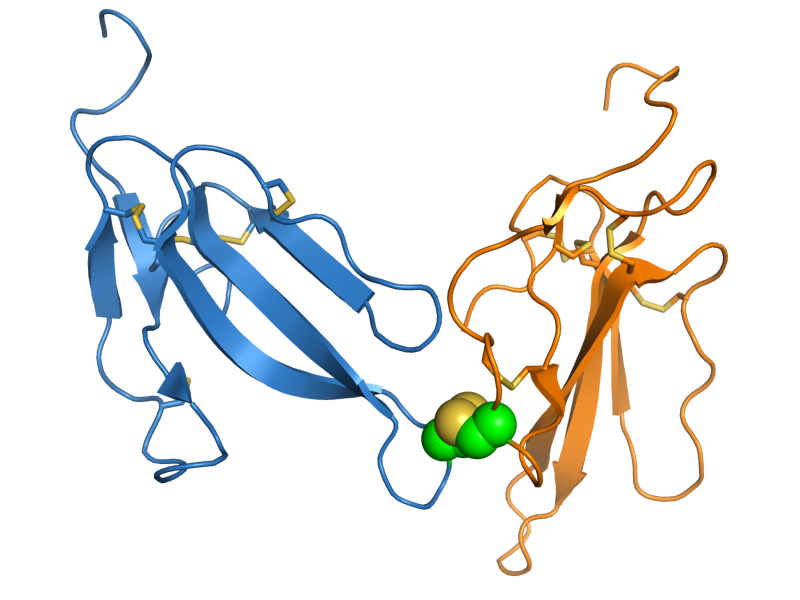
Irditoxin
Irditoxin is a three-finger toxin (3FTx) protein found in the venom of the brown tree snake (''Boiga irregularis'') and likely in other members of the genus ''Boiga''. It is a heterodimer composed of two distinct protein chains, each of the three-finger protein fold, linked by an intermolecular disulfide bond. This structure is unusual for 3FTx proteins, which are most commonly monomeric. Structure Three-finger toxin (3FTx) proteins canonically consist of approximately 60-80 amino acid residues that assume a structure with three "finger"-like beta strand-containing loops projecting from a core stabilized by four intramolecular disulfide bonds. Irditoxin is a covalent heterodimer in which two subunits are linked by an intermolecular disulfide bond. Each subunit is of the three-finger toxin (3FTx) protein superfamily and is most closely related to the "non-conventional" 3FTx subclass, characterized by the presence of an additional disulfide bond in the first of the canonical th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminal
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a protein often cont ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that j ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrophobic
In chemistry, hydrophobicity is the physical property of a molecule that is seemingly repelled from a mass of water (known as a hydrophobe). In contrast, hydrophiles are attracted to water. Hydrophobic molecules tend to be nonpolar and, thus, prefer other neutral molecules and nonpolar solvents. Because water molecules are polar, hydrophobes do not dissolve well among them. Hydrophobic molecules in water often cluster together, forming micelles. Water on hydrophobic surfaces will exhibit a high contact angle. Examples of hydrophobic molecules include the alkanes, oils, fats, and greasy substances in general. Hydrophobic materials are used for oil removal from water, the management of oil spills, and chemical separation processes to remove non-polar substances from polar compounds. Hydrophobic is often used interchangeably with lipophilic, "fat-loving". However, the two terms are not synonymous. While hydrophobic substances are usually lipophilic, there are exceptions, suc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tertiary Structure
Protein tertiary structure is the three dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure.Branden C. and Tooze J. "Introduction to Protein Structure" Garland Publishing, New York. 1990 and 1991. A number of tertiary structures may fold into a quaternary structure.Kyte, J. "Structure in Protein Chemistry." Garland Publishing, New York. 1995. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypept ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |