|
Tetraparvovirus
Tetraparvovirus are a genus of viruses in the family ''Parvoviridae''. There are six recognized species: '' Chiropteran tetraparvovirus 1'', '' Primate tetraparvovirus 1'', '' Ungulate tetraparvovirus 1'', '' Ungulate tetraparvovirus 2'', '' Ungulate tetraparvovirus 3'', and '' Ungulate tetraparvovirus 4''. History The first member of this genus was identified in 2001 in pig serum and designated Porcine parvovirus 2.Hijikata M, Abe K, Win KM, Shimizu YK, Keicho N, ''et al'' (2001) Identification of new parvovirus DNA sequence in swine sera from Myanmar. Jpn J. Infect. Dis 54: 244-245. The first human tetraparvovirus, PARV4, was described in 2005.Jones MS, Kapoor A, Lukashov VV, Simmonds P, Hecht F, ''et al'' (2005) New DNA viruses identified in patients with acute viral infection syndrome. J Virol 79: 8230ŌĆō8236. These new viruses were recognised as being related to but distinct from the known parvoviruses. They were isolated from a group of patients who had engaged in high risk ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Parvovirinae
''Parvovirinae'' is a subfamily of viruses in the family ''Parvoviridae''. There are ten genera and 84 species assigned to this subfamily. Taxonomy The following 10 genera are recognized: *'' Amdoparvovirus'' *'' Artiparvovirus'' *'' Aveparvovirus'' *'' Bocaparvovirus'' *'' Copiparvovirus'' *''Dependoparvovirus'' *'' Erythroparvovirus'' *'' Loriparvovirus'' *''Protoparvovirus'' *''Tetraparvovirus Tetraparvovirus are a genus of viruses in the family ''Parvoviridae''. There are six recognized species: '' Chiropteran tetraparvovirus 1'', '' Primate tetraparvovirus 1'', '' Ungulate tetraparvovirus 1'', '' Ungulate tetraparvovirus 2'', '' Ungu ...'' References External links ICTV ''Parvovirinae'' {{Taxonbar, from=Q2055184 Parvoviruses Virus subfamilies ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Parvoviridae
Parvoviruses are a family of animal viruses that constitute the family ''Parvoviridae''. They have linear, single-stranded DNA (ssDNA) genomes that typically contain two genes encoding for a replication initiator protein, called NS1, and the protein the viral capsid is made of. The coding portion of the genome is flanked by telomeres at each end that form into hairpin loops that are important during replication. Parvovirus virions are small compared to most viruses, at 23ŌĆō28 nanometers in diameter, and contain the genome enclosed in an icosahedral capsid that has a rugged surface. Parvoviruses enter a host cell by endocytosis, travelling to the nucleus where they wait until the cell enters its replication stage. At that point, the genome is uncoated and the coding portion is replicated. Viral messenger RNA (mRNA) is then transcribed and translated, resulting in NS1 initiating replication. During replication, the hairpins repeatedly unfold, are replicated, and refold to chang ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
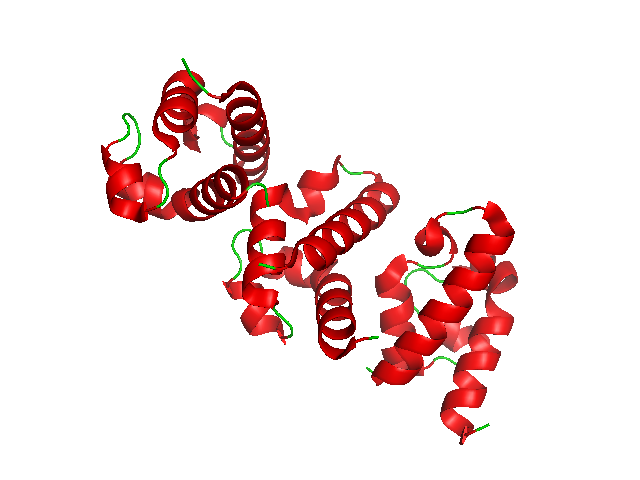
Primate Tetraparvovirus 1
Primates are a diverse order of mammals. They are divided into the strepsirrhines, which include the lemurs, galagos, and lorisids, and the haplorhines, which include the tarsiers and the simians ( monkeys and apes, the latter including humans). Primates arose 85ŌĆō55 million years ago first from small terrestrial mammals, which adapted to living in the trees of tropical forests: many primate characteristics represent adaptations to life in this challenging environment, including large brains, visual acuity, color vision, a shoulder girdle allowing a large degree of movement in the shoulder joint, and dextrous hands. Primates range in size from Madame Berthe's mouse lemur, which weighs , to the eastern gorilla, weighing over . There are 376ŌĆō524 species of living primates, depending on which classification is used. New primate species continue to be discovered: over 25 species were described in the 2000s, 36 in the 2010s, and three in the 2020s. Primates have large ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ungulate Tetraparvovirus 1
Ungulates ( ) are members of the diverse clade Ungulata which primarily consists of large mammals with hooves. These include odd-toed ungulates such as horses, rhinoceroses, and tapirs; and even-toed ungulates such as cattle, pigs, giraffes, camels, sheep, deer, and hippopotamuses. Cetaceans such as whales, dolphins, and porpoises are also classified as even-toed ungulates, although they do not have hooves. Most terrestrial ungulates use the hoofed tips of their toes to support their body weight while standing or moving. The term means, roughly, "being hoofed" or "hoofed animal". As a descriptive term, "ungulate" normally excludes cetaceans as they do not possess most of the typical morphological characteristics of other ungulates, but recent discoveries indicate that they were also descended from early artiodactyls. Ungulates are typically herbivorous and many employ specialized gut bacteria to allow them to digest cellulose. Some modern species, such as pigs, are omnivorou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ungulate Tetraparvovirus 2
Ungulates ( ) are members of the diverse clade Ungulata which primarily consists of large mammals with hooves. These include odd-toed ungulates such as horses, rhinoceroses, and tapirs; and even-toed ungulates such as cattle, pigs, giraffes, camels, sheep, deer, and hippopotamuses. Cetaceans such as whales, dolphins, and porpoises are also classified as even-toed ungulates, although they do not have hooves. Most terrestrial ungulates use the hoofed tips of their toes to support their body weight while standing or moving. The term means, roughly, "being hoofed" or "hoofed animal". As a descriptive term, "ungulate" normally excludes cetaceans as they do not possess most of the typical morphological characteristics of other ungulates, but recent discoveries indicate that they were also descended from early artiodactyls. Ungulates are typically herbivorous and many employ specialized gut bacteria to allow them to digest cellulose. Some modern species, such as pigs, ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ungulate Tetraparvovirus 3
Ungulates ( ) are members of the diverse clade Ungulata which primarily consists of large mammals with hooves. These include odd-toed ungulates such as horses, rhinoceroses, and tapirs; and even-toed ungulates such as cattle, pigs, giraffes, camels, sheep, deer, and hippopotamuses. Cetaceans such as whales, dolphins, and porpoises are also classified as even-toed ungulates, although they do not have hooves. Most terrestrial ungulates use the hoofed tips of their toes to support their body weight while standing or moving. The term means, roughly, "being hoofed" or "hoofed animal". As a descriptive term, "ungulate" normally excludes cetaceans as they do not possess most of the typical morphological characteristics of other ungulates, but recent discoveries indicate that they were also descended from early artiodactyls. Ungulates are typically herbivorous and many employ specialized gut bacteria to allow them to digest cellulose. Some modern species, such as pigs, ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ungulate Tetraparvovirus 4
Ungulates ( ) are members of the diverse clade Ungulata which primarily consists of large mammals with hooves. These include odd-toed ungulates such as horses, rhinoceroses, and tapirs; and even-toed ungulates such as cattle, pigs, giraffes, camels, sheep, deer, and hippopotamuses. Cetaceans such as whales, dolphins, and porpoises are also classified as even-toed ungulates, although they do not have hooves. Most terrestrial ungulates use the hoofed tips of their toes to support their body weight while standing or moving. The term means, roughly, "being hoofed" or "hoofed animal". As a descriptive term, "ungulate" normally excludes cetaceans as they do not possess most of the typical morphological characteristics of other ungulates, but recent discoveries indicate that they were also descended from early artiodactyls. Ungulates are typically herbivorous and many employ specialized gut bacteria to allow them to digest cellulose. Some modern species, such as pigs, ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an ORF may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to yield the final ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helicase
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands (hence '' helic- + -ase''), using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases. The human genome codes for 95 non-redundant helicases: 64 RNA helicases and 31 DNA helicases. Many cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair, and ribosome biogenesis involve the separation of nucleic acid strands that necessitates the use of helicases. Some specialized helicases are also involved in sensing of viral nucleic acids during infection and fulfill a immunological function. Function Helicases are o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATPase
ATPases (, Adenosine 5'-TriPhosphatase, adenylpyrophosphatase, ATP monophosphatase, triphosphatase, SV40 T-antigen, ATP hydrolase, complex V (mitochondrial electron transport), (Ca2+ + Mg2+)-ATPase, HCO3ŌłÆ-ATPase, adenosine triphosphatase) are a class of enzymes that catalyze the decomposition of ATP into ADP and a free phosphate ion or the inverse reaction. This dephosphorylation reaction releases energy, which the enzyme (in most cases) harnesses to drive other chemical reactions that would not otherwise occur. This process is widely used in all known forms of life. Some such enzymes are integral membrane proteins (anchored within biological membranes), and move solutes across the membrane, typically against their concentration gradient. These are called transmembrane ATPases. Functions Transmembrane ATPases import metabolites necessary for cell metabolism and export toxins, wastes, and solutes that can hinder cellular processes. An important example is the sodium-potass ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phospholipase
A phospholipase is an enzyme that hydrolyzes phospholipids into fatty acids and other lipophilic substances. Acids trigger the release of bound calcium from cellular stores and the consequent increase in free cytosolic Ca2+, an essential step in calcium signaling to regulate intracellular processes. There are four major classes, termed A, B, C, and D, which are distinguished by the type of reaction which they catalyze: *Phospholipase A **Phospholipase A1 ŌĆō cleaves the ''sn''-1 acyl chain (where ''sn'' refers to stereospecific numbering). **Phospholipase A2 ŌĆō cleaves the ''sn''-2 acyl chain, releasing arachidonic acid. * Phospholipase B ŌĆō cleaves both ''sn''-1 and ''sn''-2 acyl chains; this enzyme is also known as a lysophospholipase. *Phospholipase C ŌĆō cleaves before the phosphate, releasing diacylglycerol and a phosphate-containing head group. PLCs play a central role in signal transduction, releasing the second messenger inositol triphosphate. * Phospholipase D ŌĆ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

.jpg)