|
Telomere-binding Protein
Telomere-binding proteins (also known as TERF, TRBF, TRF) function to bind telomeric DNA in various species. In particular, telomere-binding protein refers to TTAGGG repeat binding factor-1 (TERF1) and TTAGGG repeat binding factor-2 (TERF2). Telomere sequences in humans are composed of TTAGGG sequences which provide protection and replication of chromosome ends to prevent degradation. Telomere-binding proteins can generate a T-loop to protect chromosome ends. TRFs are double-stranded proteins which are known to induce bending, looping, and pairing of DNA which aids in the formation of T-loops. They directly bind to TTAGGG repeat sequence in the DNA. There are also subtelomeric regions present for regulation. However, in humans, there are six subunits forming a complex known as shelterin. Structure There are six subunits forming the telomere-binding protein complex known as shelterin: TERF1, TERF2, POT1, TIN2, RAP1 and TPP1. Both TERF1 and TERF2 bind the telomeric repeat seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telomere
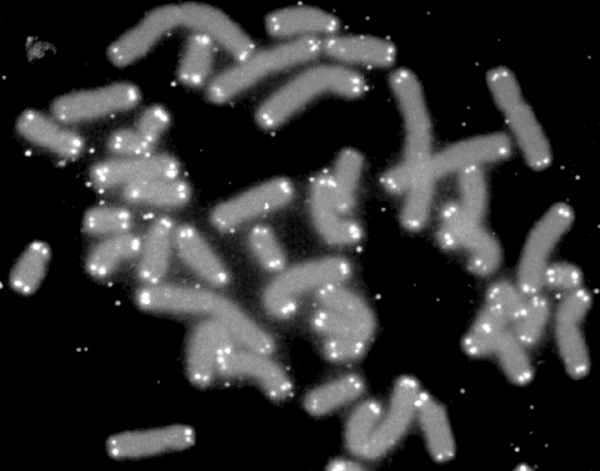
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes. Although there are different architectures, telomeres, in a broad sense, are a widespread genetic feature most commonly found in eukaryotes. In most, if not all species possessing them, they protect the terminal regions of chromosomal DNA from progressive degradation and ensure the integrity of linear chromosomes by preventing DNA repair systems from mistaking the very ends of the DNA strand for a double-strand break. Discovery In the early 1970s, Soviet theorist Alexei Olovnikov first recognized that chromosomes could not completely replicate their ends; this is known as the "end replication problem". Building on this, and accommodating Leonard Hayflick's idea of limited somatic cell division, Olovnikov suggested that DNA sequences are lost every time a cell replicates until the loss reaches a critical level, at which point cell division end ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helicase
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands (hence '' helic- + -ase''), using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases. The human genome codes for 95 non-redundant helicases: 64 RNA helicases and 31 DNA helicases. Many cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair, and ribosome biogenesis involve the separation of nucleic acid strands that necessitates the use of helicases. Some specialized helicases are also involved in sensing of viral nucleic acids during infection and fulfill a immunological function. Function Helicases are o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Xeroderma Pigmentosum
Xeroderma pigmentosum (XP) is a genetic disorder in which there is a decreased ability to repair DNA damage such as that caused by ultraviolet (UV) light. Symptoms may include a severe sunburn after only a few minutes in the sun, freckling in sun-exposed areas, dry skin and changes in skin pigmentation. Nervous system problems, such as hearing loss, poor coordination, loss of intellectual function and seizures, may also occur. Complications include a high risk of skin cancer, with about half having skin cancer by age 10 without preventative efforts, and cataracts. There may be a higher risk of other cancers such as brain cancers. XP is autosomal recessive, with mutations in at least nine specific genes able to result in the condition. Normally, the damage to DNA which occurs in skin cells from exposure to UV light is repaired by nucleotide excision repair. In people with xeroderma pigmentosum, this damage is not repaired. As more abnormalities form in DNA, cells malfunction and e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide Excision Repair
Nucleotide excision repair is a DNA repair mechanism. DNA damage occurs constantly because of chemicals (e.g. intercalating agents), radiation and other mutagens. Three excision repair pathways exist to repair single stranded DNA damage: Nucleotide excision repair (NER), base excision repair (BER), and DNA mismatch repair (MMR). While the BER pathway can recognize specific non-bulky lesions in DNA, it can correct only damaged bases that are removed by specific glycosylases. Similarly, the MMR pathway only targets mismatched Watson-Crick base pairs. Nucleotide excision repair (NER) is a particularly important excision mechanism that removes DNA damage induced by ultraviolet light (UV). UV DNA damage results in bulky DNA adducts - these adducts are mostly thymine dimers and 6,4-photoproducts. Recognition of the damage leads to removal of a short single-stranded DNA segment that contains the lesion. The undamaged single-stranded DNA remains and DNA polymerase uses it as a templa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-homologous End Joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair(HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber. NHEJ is typically guided by short homologous DNA sequences called microhomologies. These microhomologies are often present in single-stranded overhangs on the ends of double-strand breaks. When the overhangs are perfectly compatible, NHEJ usually repairs the break accurately. Imprecise repair leading to loss of nucleotides can also occur, but is much more common when the overhangs are not compatible. Inappropriate NHEJ can lead to translocations and telomere fusion, hallmarks ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apoptosis
Apoptosis (from grc, ἀπόπτωσις, apóptōsis, 'falling off') is a form of programmed cell death that occurs in multicellular organisms. Biochemical events lead to characteristic cell changes (morphology) and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, DNA fragmentation, and mRNA decay. The average adult human loses between 50 and 70 billion cells each day due to apoptosis. For an average human child between eight and fourteen years old, approximately twenty to thirty billion cells die per day. In contrast to necrosis, which is a form of traumatic cell death that results from acute cellular injury, apoptosis is a highly regulated and controlled process that confers advantages during an organism's life cycle. For example, the separation of fingers and toes in a developing human embryo occurs because cells between the digits undergo apoptosis. Unlike necrosis, apoptosis produces cell fragments called apoptotic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TP53
p53, also known as Tumor protein P53, cellular tumor antigen p53 (UniProt name), or transformation-related protein 53 (TRP53) is a regulatory protein that is often mutated in human cancers. The p53 proteins (originally thought to be, and often spoken of as, a single protein) are crucial in vertebrates, where they prevent cancer formation. As such, p53 has been described as "the guardian of the genome" because of its role in conserving stability by preventing genome mutation. Hence ''TP53'' ''italics'' are used to denote the ''TP53'' gene name and distinguish it from the protein it encodes is classified as a tumor suppressor gene. The name p53 was given in 1979 describing the apparent molecular mass; SDS-PAGE analysis indicates that it is a 53-kilodalton (kDa) protein. However, the actual mass of the full-length p53 protein (p53α) based on the sum of masses of the amino acid residues is only 43.7 kDa. This difference is due to the high number of proline residues in the protein, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TP53BP1
Tumor suppressor p53-binding protein 1 also known as p53-binding protein 1 or 53BP1 is a protein that in humans is encoded by the ''TP53BP1'' gene. Clinical significance 53BP1 is underexpressed in most cases of triple-negative breast cancer. DNA repair DNA double-strand breaks (DSBs) are cytotoxic damages that can be repaired either by the homologous recombinational repair (HR) pathway or by the non-homologous end-joining (NHEJ) pathway. NHEJ, although faster than HR, is less accurate. The early divergent step between the two pathways is end resection, and this step is regulated by numerous factors. In particular, BRCA1 and 53BP1 play a role in determining the balance between the two pathways. 53BP1 restricts resection and promotes NHEJ. Age-associated deficient repair Ordinarily during the G1 phase of the cell cycle, when a sister chromatid is unavailable for HR, NHEJ is the predominant pathway for repairing DNA double-strand breaks (DSBs). However, as individuals age, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H2AFX
H2A histone family member X (usually abbreviated as H2AX) is a type of histone protein from the H2A family encoded by the ''H2AFX'' gene. An important phosphorylated form is γH2AX (S139), which forms when double-strand breaks appear. In humans and other eukaryotes, the DNA is wrapped around histone octamers, consisting of core histones H2A, H2B, H3 and H4, to form chromatin. H2AX contributes to nucleosome-formation, chromatin-remodeling and DNA repair, and is also used ''in vitro'' as an assay for double-strand breaks in dsDNA. Formation of γH2AX H2AX becomes phosphorylated on serine 139, then called γH2AX, as a reaction on DNA double-strand breaks (DSB). The kinases of the PI3-family ( Ataxia telangiectasia mutated, ATR and DNA-PKcs) are responsible for this phosphorylation, especially ATM. The modification can happen accidentally during replication fork collapse or in the response to ionizing radiation but also during controlled physiological processes such as V(D)J ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages (interstrand crosslinks or ICLs). This can eventually lead to malignant ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATM Serine/threonine Kinase
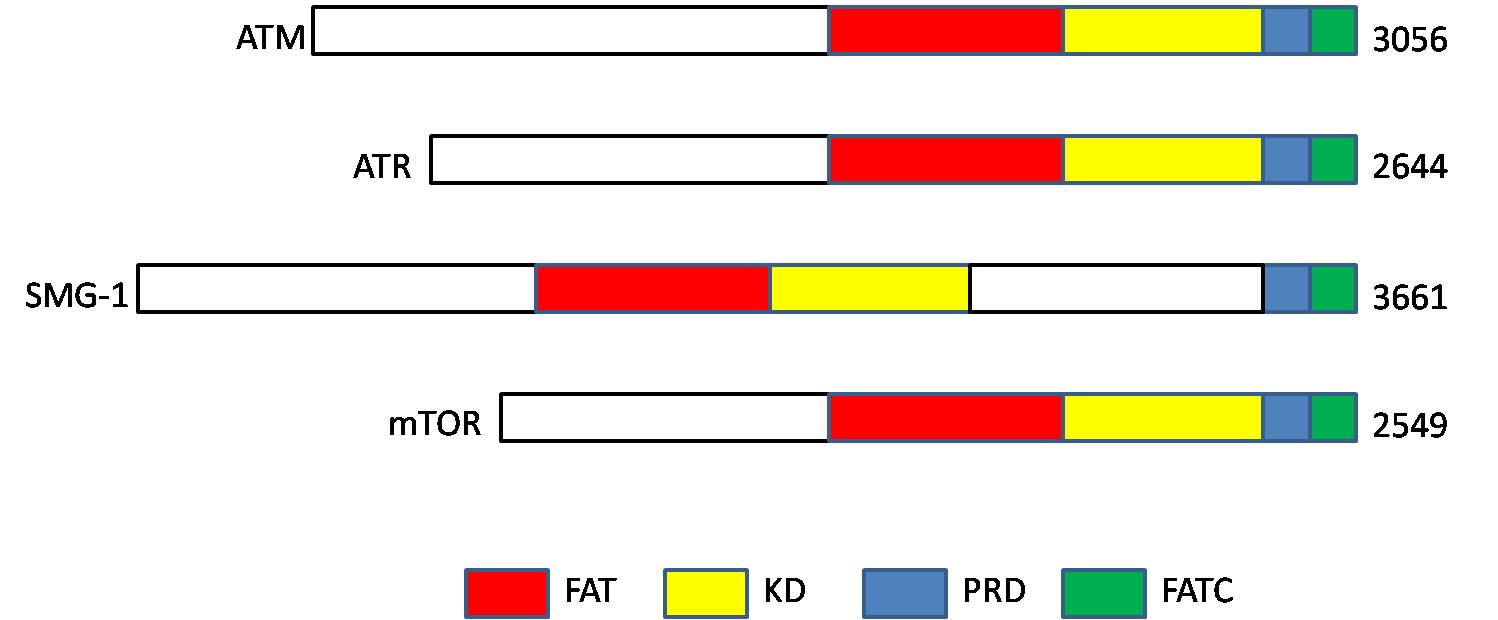
ATM serine/threonine kinase or Ataxia-telangiectasia mutated, symbol ATM, is a serine/ threonine protein kinase that is recruited and activated by DNA double-strand breaks. It phosphorylates several key proteins that initiate activation of the DNA damage checkpoint, leading to cell cycle arrest, DNA repair or apoptosis. Several of these targets, including p53, CHK2, BRCA1, NBS1 and H2AX are tumor suppressors. In 1995, the gene was discovered by Yosef Shiloh who named its product ATM since he found that its mutations are responsible for the disorder ataxia–telangiectasia. In 1998, the Shiloh and Kastan laboratories independently showed that ATM is a protein kinase whose activity is enhanced by DNA damage. Introduction Throughout the cell cycle DNA is monitored for damage. Damages result from errors during replication, by-products of metabolism, general toxic drugs or ionizing radiation. The cell cycle has different DNA damage checkpoints, which inhibit the next or maint ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ERCC4
ERCC4 is a protein designated as DNA repair endonuclease XPF that in humans is encoded by the ''ERCC4'' gene. Together with ERCC1, ERCC4 forms the ERCC1-XPF enzyme complex that participates in DNA repair and DNA recombination. The nuclease enzyme ERCC1-XPF cuts specific structures of DNA. Many aspects of these two gene products are described together here because they are partners during DNA repair. The ERCC1-XPF nuclease is an essential activity in the pathway of DNA nucleotide excision repair (NER). The ERCC1-XPF nuclease also functions in pathways to repair double-strand breaks in DNA, and in the repair of "crosslink" damage that harmfully links the two DNA strands. Cells with disabling mutations in ''ERCC4'' are more sensitive than normal to particular DNA damaging agents, including ultraviolet radiation and to chemicals that cause crosslinking between DNA strands. Genetically engineered mice with disabling mutations in ''ERCC4'' also have defects in DNA repair, accompanie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |