|
TALE-likes
Transcription Activator-Like Effector-Likes (TALE-likes) are a group of bacterial DNA binding proteins named for the first and still best-studied group, the TALEs of ''Xanthomonas'' bacteria. TALEs are important factors in the plant diseases caused by ''Xanthomonas'' bacteria, but are known primarily for their role in biotechnology as programmable DNA binding proteins, particularly in the context of TALE nucleases. TALE-likes have additionally been found in many strains of the ''Ralstonia solanacearum'' bacterial species complex, in ''Paraburkholderia rhizoxinica'' strain HKI 454, and in two unknown marine bacteria. Whether or not all these proteins form a single phylogenetic grouping is as yet unclear. The unifying feature of the TALE-likes are their tandem arrays of DNA binding repeats. These repeats are, with few exceptions, 33-35 amino acids in length, and composed of two alpha-helices on either side of a flexible loop containing the DNA base binding residues and with nei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TAL Effector
TAL (transcription activator-like) effectors (often referred to as TALEs, but not to be confused with the three amino acid loop extension homeobox class of proteins) are proteins secreted by some β- and γ-proteobacteria. Most of these are Xanthomonads. Plant pathogenic ''Xanthomonas'' bacteria are especially known for TALEs, produced via their type III secretion system. These proteins can bind promoter sequences in the host plant and activate the expression of plant genes that aid bacterial infection. The TALE domain responsible for binding to DNA is known to have 1.5 to 33.5 short sequences that are repeated multiple times (tandem repeats). Each of these repeats was found to be specific for a certain base pair of the DNA. These repeats also have repeat variable residues (RVD) that can detect specific DNA base pairs. They recognize plant DNA sequences through a central repeat domain consisting of a variable number of ~34 amino acid repeats. There appears to be a one-to-one ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Logo
In bioinformatics, a sequence logo is a graphical representation of the sequence conservation of nucleotides (in a strand of DNA/RNA) or amino acids (in protein sequences). A sequence logo is created from a collection of aligned sequences and depicts the consensus sequence and diversity of the sequences. Sequence logos are frequently used to depict sequence characteristics such as protein-binding sites in DNA or functional units in proteins. Overview A sequence logo consists of a stack of letters at each position. The relative sizes of the letters indicate their frequency in the sequences. The total height of the letters depicts the information content of the position, in bits. Logo creation To create sequence logos, related DNA, RNA or protein sequences, or DNA sequences that have common conserved binding sites, are aligned so that the most conserved parts create good alignments. A sequence logo can then be created from the conserved multiple sequence alignment. The sequence ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA-binding Protein
DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for DNA#Base pairing, single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair. However, there are some known minor groove DNA-binding ligands such as netropsin, distamycin, Hoechst 33258, pentamidine, DAPI and others. Examples DNA-binding proteins include transcription factors which Gene modulation, modulate the process of transcription, various polymerases, nucleases which cleave DNA molecules, and histones which are involved in chromosome packaging and transcription in the cell nucleus. DNA-binding proteins can incorporate such domains as the zinc finger, the helix-turn-helix, and the leucine zipper (among many others) that facilitate binding to nucleic acid. There are also more unusual examples such as TAL effector, transcription activa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Activator-like Effector Nuclease
Transcription activator-like effector nucleases (TALEN) are restriction enzymes that can be engineered to cut specific sequences of DNA. They are made by fusing a TAL effector DNA-binding domain to a DNA cleavage domain (a nuclease which cuts DNA strands). Transcription activator-like effectors (TALEs) can be engineered to bind to practically any desired DNA sequence, so when combined with a nuclease, DNA can be cut at specific locations. The restriction enzymes can be introduced into cells, for use in gene editing or for genome editing ''in situ'', a technique known as genome editing with engineered nucleases. Alongside zinc finger nucleases and CRISPR/Cas9, TALEN is a prominent tool in the field of genome editing. TALE DNA-binding domain TAL effectors are proteins that are secreted by ''Xanthomonas'' bacteria via their type III secretion system when they infect plants. The DNA binding domain contains a repeated highly conserved 33–34 amino acid sequence with divergent 12th a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BATs
Bats are mammals of the order Chiroptera.''cheir'', "hand" and πτερόν''pteron'', "wing". With their forelimbs adapted as wings, they are the only mammals capable of true and sustained flight. Bats are more agile in flight than most birds, flying with their very long spread-out digits covered with a thin membrane or patagium. The smallest bat, and arguably the smallest extant mammal, is Kitti's hog-nosed bat, which is in length, across the wings and in mass. The largest bats are the flying foxes, with the giant golden-crowned flying fox, ''Acerodon jubatus'', reaching a weight of and having a wingspan of . The second largest order of mammals after rodents, bats comprise about 20% of all classified mammal species worldwide, with over 1,400 species. These were traditionally divided into two suborders: the largely fruit-eating megabats, and the echolocating microbats. But more recent evidence has supported dividing the order into Yinpterochiroptera and Yangochiropte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ralstonia Solanacearum
''Ralstonia solanacearum'' is an aerobic non-spore-forming, Gram-negative, plant pathogenic bacterium. ''R. solanacearum'' is soil-borne and motile with a polar flagellar tuft. It colonises the xylem, causing bacterial wilt in a very wide range of potential host plants. It is known as Granville wilt when it occurs in tobacco. Bacterial wilts of tomato, pepper, eggplant, and Irish potato caused by ''R. solanacearum'' were among the first diseases that Erwin Frink Smith proved to be caused by a bacterial pathogen. Because of its devastating lethality, ''R. solanacearum'' is now one of the more intensively studied phytopathogenic bacteria, and bacterial wilt of tomato is a model system for investigating mechanisms of pathogenesis. ''Ralstonia'' was until recently classified as ''Pseudomonas'', with similarity in most aspects, except that it does not produce fluorescent pigment like ''Pseudomonas''. The genomes from different strains vary from 5.5 Mb up to 6 Mb, roughly being 3.5 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Type Three Secretion System
Type three secretion system (often written Type III secretion system and abbreviated TTSS or T3SS, also called Injectisome) is a protein appendage found in several Gram-negative bacteria. In pathogenic bacteria, the needle-like structure is used as a sensory probe to detect the presence of eukaryotic organisms and secrete proteins that help the bacteria infect them. The secreted effector proteins are secreted directly from the bacterial cell into the eukaryotic (host) cell, where they exert a number of effects that help the pathogen to survive and to escape an immune response. Overview The term Type III secretion system was coined in 1993. This secretion system is distinguished from at least five other secretion systems found in Gram-negative bacteria. Many animal and plant associated bacteria possess similar T3SSs. These T3SSs are similar as a result of divergent evolution and phylogenetic analysis supports a model in which gram-negative bacteria can transfer the T3SS gene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paraburkholderia Rhizoxinica
''Paraburkholderia rhizoxinica'' is a gram-negative, oxidase and catalase-positiv, motile bacterium from the genus ''Paraburkholderia'' and the family Burkholderiaceae which was isolated from the plant pathogenic fungus, ''Rhizopus microsporus''. The complete genome of ''Paraburkholderia rhizoxinica'' is sequenced In genetics and biochemistry, sequencing means to determine the primary structure (sometimes incorrectly called the primary sequence) of an unbranched biopolymer. Sequencing results in a symbolic linear depiction known as a sequence which suc .... References rhizoxinica Bacteria described in 2007 {{Betaproteobacteria-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FokI
The restriction endonuclease ''Fok''1, naturally found in ''Flavobacterium okeanokoites'', is a bacterial type IIS restriction endonuclease consisting of an N-terminal DNA-binding domain and a non-specific DNA cleavage domain at the C-terminal. Once the protein is bound to duplex DNA via its DNA-binding domain at the 5'-GGATG-3' recognition site, the DNA cleavage domain is activated and cleaves, without further sequence specificity, the first strand 9 nucleotides downstream and the second strand 13 nucleotides upstream of the nearest nucleotide of the recognition site. Its molecular mass is 65.4 kDa, being composed of 587 amino acids. DNA-binding domain The recognition domain contains three subdomains (D1, D2 and D3) that are evolutionarily related to the DNA-binding domain of the catabolite gene activator protein which contains a helix-turn-helix. DNA-cleavage domain DNA cleavage is mediated through the non-specific cleavage domain which also includes the dimerisation su ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
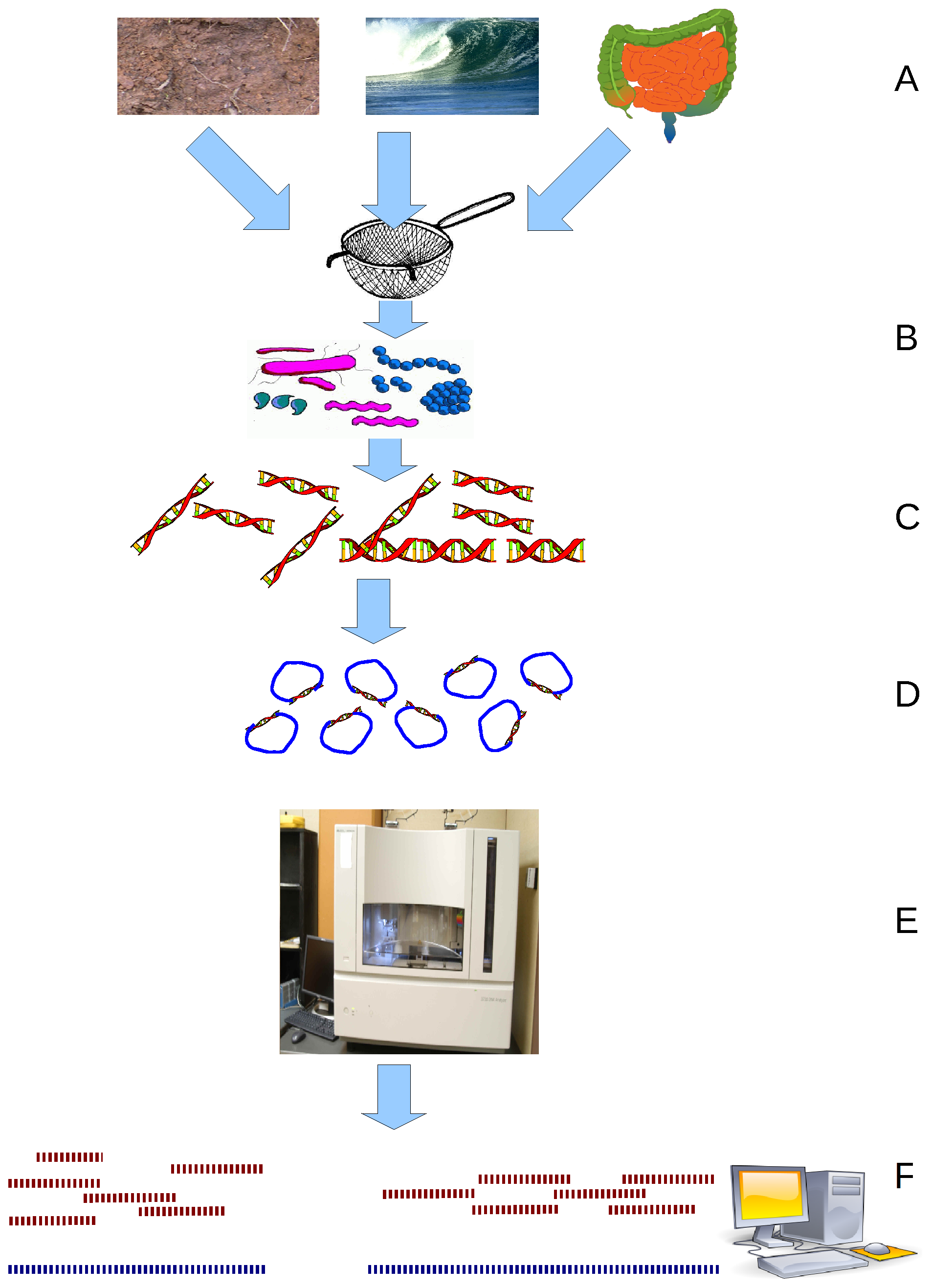
Metagenomic
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |