|
T-bet
T-box transcription factor TBX21, also called T-bet (T-box expressed in T cells) is a protein that in humans is encoded by the ''TBX21'' gene. Though being for long thought of only as a master regulator of type 1 immune response, T-bet has recently been shown to be implicated in development of various immune cell subsets and maintenance of mucosal homeostasis. Function This gene is a member of a phylogenetically conserved family of genes that share a common DNA-binding domain, the T-box. T-box genes encode transcription factors involved in the regulation of developmental processes. This gene is the human ortholog of mouse Tbx21/Tbet gene. Studies in mouse show that Tbx21 protein is a Th1 cell-specific transcription factor that controls the expression of the hallmark Th1 cytokine, interferon-gamma ( IFNg). Expression of the human ortholog also correlates with IFNg expression in Th1 and natural killer cells, suggesting a role for this gene in initiating Th1 lineage development fro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
STAT1
Signal transducer and activator of transcription 1 (STAT1) is a transcription factor which in humans is encoded by the ''STAT1'' gene. It is a member of the STAT protein family. Function All STAT molecules are phosphorylated by receptor associated kinases, that causes activation, dimerization by forming homo- or heterodimers and finally translocate to nucleus to work as transcription factors. Specifically STAT1 can be activated by several ligands such as Interferon alpha (IFNα), Interferon gamma (IFNγ), Epidermal Growth Factor (EGF), Platelet Derived Growth Factor (PDGF), Interleukin 6 (IL-6), or IL-27. Type I interferons (IFN-α, IFN-ß) bind to receptors, cause signaling via kinases, phosphorylate and activate the Jak kinases TYK2 and JAK1 and also STAT1 and STAT2. STAT molecules form dimers and bind to ISGF3G/IRF-9, which is Interferon stimulated gene factor 3 complex with Interferon regulatory Factor 9. This allows STAT1 to enter the nucleus. STAT1 has a key role in many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Th1 Cell
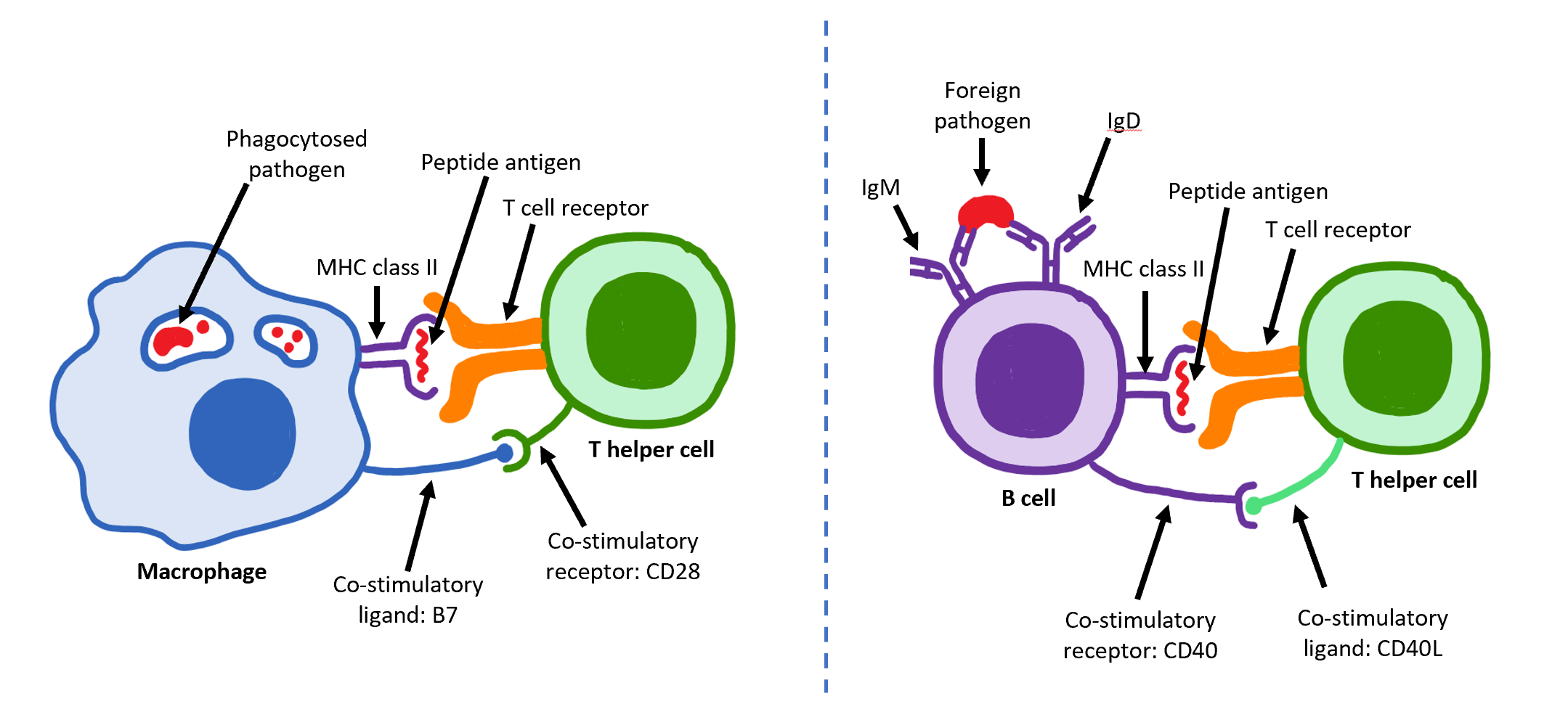
The T helper cells (Th cells), also known as CD4+ cells or CD4-positive cells, are a type of T cell that play an important role in the adaptive immune system. They aid the activity of other immune cells by releasing cytokines. They are considered essential in B cell antibody class switching, breaking cross-tolerance in dendritic cells, in the activation and growth of cytotoxic T cells, and in maximizing bactericidal activity of phagocytes such as macrophages and neutrophils. CD4+ cells are mature Th cells that express the surface protein CD4. Genetic variation in regulatory elements expressed by CD4+ cells determines susceptibility to a broad class of autoimmune diseases. Structure and function Th cells contain and release cytokines to aid other immune cells. Cytokines are small protein mediators that alter the behavior of target cells that express receptors for those cytokines. These cells help polarize the immune response depending on the nature of the immunological insul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
T Helper Cell
The T helper cells (Th cells), also known as CD4+ cells or CD4-positive cells, are a type of T cell that play an important role in the adaptive immune system. They aid the activity of other immune cells by releasing cytokines. They are considered essential in B cell antibody class switching, breaking cross-tolerance in dendritic cells, in the activation and growth of cytotoxic T cells, and in maximizing bactericidal activity of phagocytes such as macrophages and neutrophils. CD4+ cells are mature Th cells that express the surface protein CD4. Genetic variation in regulatory elements expressed by CD4+ cells determines susceptibility to a broad class of autoimmune diseases. Structure and function Th cells contain and release cytokines to aid other immune cells. Cytokines are small protein mediators that alter the behavior of target cells that express receptors for those cytokines. These cells help polarize the immune response depending on the nature of the immunological in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GATA3
GATA3 is a transcription factor that in humans is encoded by the ''GATA3'' gene. Studies in animal models and humans indicate that it controls the expression of a wide range of biologically and clinically important genes. The GATA3 transcription factor is critical for the embryonic development of various tissues as well as for inflammatory and humoral immune responses and the proper functioning of the endothelium of blood vessels. GATA3 plays central role in allergy and immunity against worm infections. ''GATA3'' haploinsufficiency (i.e. loss of one or the two inherited ''GATA3'' genes) results in a congenital disorder termed the Barakat syndrome. Current clinical and laboratory research is focusing on determining the benefits of directly or indirectly blocking the action of GATA3 in inflammatory and allergic diseases such as asthma. It is also proposed to be a clinically important marker for various types of cancer, particularly those of the breast. However, the role, if any, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enhancer (genetics)
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are ''cis''-acting. They can be located up to 1 Mbp (1,000,000 bp) away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes. The first discovery of a eukaryotic enhancer was in the immunoglobulin heavy chain gene in 1983. This enhancer, located in the large intron, provided an explanation for the transcriptional activation of rearranged Vh gene promoters while unrearranged Vh promoters remained inactive. Locations In eukaryotic cells the structure of the chromatin complex of DNA is folded in a way that functionally mimics the supercoiled state characteristic of prokaryotic DNA, so although the en ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HLX (gene)
Homeobox Protein HB24 is a protein that in humans is encoded by the ''HLX'' gene. Role in development Hlx belongs to the class of homeobox transcription factors, initially cloned from a B-lymphocyte cell line. Targeted knockout of the gene has demonstrated its vital role in liver and gut organogenesis. Its expression is first noticed in embryonic day 9.5 (E9.5) in the splanchnic mesoderm caudal to the level of the heart and foregut pocket, and in the branchial arches. Around E10- E12.5, the expression becomes more prominent in the mesenchyme of the visceral organs of the gut such as liver, intestines and gall bladder. Hlx is essential for liver and gut expansion, but not for onset of their development. Heterozygous knockouts of Hlx (Hlx +/−) are normal whereas homozygous knockouts (Hlx −/–) develop severe hypoplasia of the liver and gut along with anaemia. Hlx controls the epithelial-mesenchymal interaction necessary for liver and gut expansion. At E8.0, the primary live ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RUNX1
Runt-related transcription factor 1 (RUNX1) also known as acute myeloid leukemia 1 protein (AML1) or core-binding factor subunit alpha-2 (CBFA2) is a protein that in humans is encoded by the ''RUNX1'' gene. RUNX1 is a transcription factor that regulates the differentiation of hematopoietic stem cells into mature blood cells. In addition it plays a major role in the development of the neurons that transmit pain. It belongs to the Runt-related transcription factor (RUNX) family of genes which are also called core binding factor-α (CBFα). RUNX proteins form a heterodimeric complex with CBFβ which confers increased DNA binding and stability to the complex. Chromosomal translocations involving the ''RUNX1'' gene are associated with several types of leukemia including M2 AML. Mutations in ''RUNX1'' are implicated in cases of breast cancer. Gene and protein In humans, the gene RUNX1 is 260 kilobases (kb) in length, and is located on chromosome 21 (21q22.12). The gene can be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RUNX3
Runt-related transcription factor 3 is a protein that in humans is encoded by the ''RUNX3'' gene. Function This gene encodes a member of the runt domain-containing family of transcription factors. A heterodimer of this protein and a beta subunit forms a complex that binds to the core DNA sequence 5'-YGYGGT-3' found in a number of enhancers and promoters, and can either activate or suppress transcription. It also interacts with other transcription factors. It functions as a tumor suppressor, and the gene is frequently deleted or transcriptionally silenced in cancer. Multiple transcript variants encoding different isoforms have been found for this gene. In melanocytic cells RUNX3 gene expression may be regulated by MITF. Knockout mouse Runx3 null mouse gastric mucosa exhibits hyperplasia due to stimulated proliferation and suppressed apoptosis in epithelial cells, and the cells are resistant to TGF-beta stimulation. The RUNX3 controversy In 2011 serious doubt was cast o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid resid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetics
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional genetic basis for inheritance. Epigenetics most often involves changes that affect the regulation of gene expression, but the term can also be used to describe any heritable phenotypic change. Such effects on cellular and physiological phenotypic traits may result from external or environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Gene expressi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ccr5
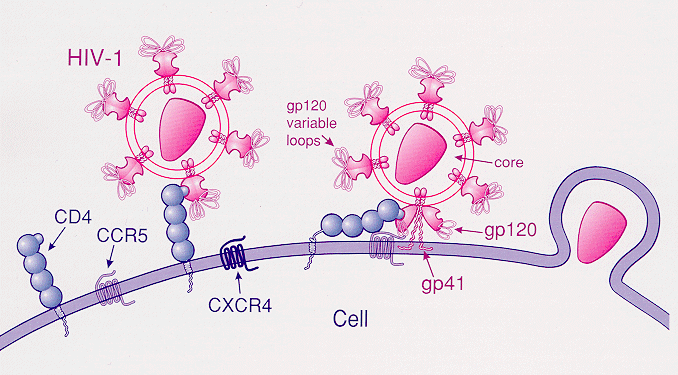
C-C chemokine receptor type 5, also known as CCR5 or CD195, is a protein on the surface of white blood cells that is involved in the immune system as it acts as a receptor for chemokines. In humans, the ''CCR5'' gene that encodes the CCR5 protein is located on the short (p) arm at position 21 on chromosome 3. Certain populations have inherited the ''Delta 32'' mutation, resulting in the genetic deletion of a portion of the CCR5 gene. Homozygous carriers of this mutation are resistant to M-tropic strains of HIV-1 infection. Function The CCR5 protein belongs to the beta chemokine receptors family of integral membrane proteins. It is a G protein–coupled receptor which functions as a chemokine receptor in the CC chemokine group. CCR5's cognate ligands include CCL3, CCL4 (also known as MIP 1''α'' and 1''β'', respectively), and CCL3L1. CCR5 furthermore interacts with CCL5 (a chemotactic cytokine protein also known as RANTES). CCR5 is predominantly expressed on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |