|
Stop Codon
In molecular biology (specifically protein biosynthesis), a stop codon (or termination codon) is a codon (nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain. While start codons need nearby sequences or initiation factors to start translation, a stop codon alone is sufficient to initiate termination. Properties Standard codons In the standard genetic code, there are three different termination codons: Alternative stop codons There are variations on the standard genetic code, and alternative stop codons have been found in the mitochondrial genomes of vertebrates, ''Scenedesmus obliquus'', and '' Thra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homo Sapiens-mtDNA~NC 012920-ATP8+ATP6 Overlap
''Homo'' () is the genus that emerged in the (otherwise extinct) genus ''Australopithecus'' that encompasses the extant species ''Homo sapiens'' ( modern humans), plus several extinct species classified as either ancestral to or closely related to modern humans (depending on the species), most notably ''Homo erectus'' and ''Homo neanderthalensis''. The genus emerged with the appearance of ''Homo habilis'' just over 2 million years ago. ''Homo'', together with the genus '' Paranthropus'', is probably sister to ''Australopithecus africanus'', which itself had previously split from the lineage of '' Pan'', the chimpanzees. ''Homo erectus'' appeared about 2 million years ago and, in several early migrations, spread throughout Africa (where it is dubbed ''Homo ergaster'') and Eurasia. It was likely that the first human species lived in a hunter-gatherer society and was able to control fire. An adaptive and successful species, ''Homo erectus'' persisted for more than a million year ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Scenedesmus Obliquus
''Scenedesmus obliquus'' is a green algae species of the genus ''Scenedesmus''. This chlorophyte species is notable for the genetic coding of its mitochondria which translate TCA as a stop codon and TAG as leucine. This code is represented by NCBI translation table 22, ''Scenedesmus obliquus'' mitochondrial code. Both growth and photosynthesis of ''S. obliquus'' are affected by the presence of nano-sized microplastics, such as nano-polystyrene (nano-PS). See also * List of genetic codes While there is much commonality, different parts of the tree of life use slightly different genetic codes. When translating from genome to protein, the use of the correct genetic code is essential. The mitochondrial codes are the relatively well-k ... References External links Sphaeropleales {{Chlorophyceae-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
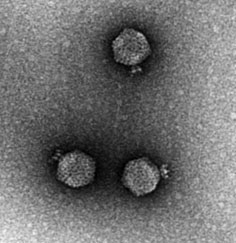
Enterobacteria Phage T4
Escherichia virus T4 is a species of bacteriophages that infect ''Escherichia coli'' bacteria. It is a double-stranded DNA virus in the subfamily '' Tevenvirinae'' from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle and not the lysogenic lifecycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains (or isolates), Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6. Use in research Dating back to the 1940s and continuing today, T-even phages are considered the best studied model organisms. Model organisms are usually required to be simple with as few as five genes. Yet, T-even phages are in fact among the largest and highest complexity virus, in which these phage's genetic information is made up of around 300 genes. Coincident with their complexity, T-even viruses were found to have the unusual base hydroxymethylcytosine (HMC) in place of the nucleic acid base cytosine. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes (e.g. MS2) and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm. Bacteriophages are among the most common and diverse entities in the biosphere. Bacteriophages are ubiquitous viruses, found wherever bacteria exist. It is estimated there are more than 1031 bacteriophages on the planet, more than every other organism on Earth, including bacteria, combined. Viruses are the most abundant biological entity in the water column of the world's oceans, and the second largest component of biom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GC-content
In molecular biology and genetics, GC-content (or guanine-cytosine content) is the percentage of nitrogenous bases in a DNA or RNA molecule that are either guanine (G) or cytosine (C). This measure indicates the proportion of G and C bases out of an implied four total bases, also including adenine and thymine in DNA and adenine and uracil in RNA. GC-content may be given for a certain fragment of DNA or RNA or for an entire genome. When it refers to a fragment, it may denote the GC-content of an individual gene or section of a gene (domain), a group of genes or gene clusters, a non-coding region, or a synthetic oligonucleotide such as a primer. Structure Qualitatively, guanine (G) and cytosine (C) undergo a specific hydrogen bonding with each other, whereas adenine (A) bonds specifically with thymine (T) in DNA and with uracil (U) in RNA. Quantitatively, each GC base pair is held together by three hydrogen bonds, while AT and AU base pairs are held together by two hydrogen bonds. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrrolysine
Pyrrolysine (symbol Pyl or O; encoded by the 'amber' stop codon UAG) is an α-amino acid that is used in the biosynthesis of proteins in some methanogenic archaea and bacteria; it is not present in humans. It contains an α-amino group (which is in the protonated – form under biological conditions), a carboxylic acid group (which is in the deprotonated –COO− form under biological conditions). Its pyrroline side-chain is similar to that of lysine in being basic and positively charged at neutral pH. Genetics Nearly all genes are translated using only 20 standard amino acid building blocks. Two unusual genetically-encoded amino acids are selenocysteine and pyrrolysine. Pyrrolysine was discovered in 2002 at the active site of methyltransferase enzyme from a methane-producing archeon, ''Methanosarcina barkeri''. This amino acid is encoded by UAG (normally a stop codon), and its synthesis and incorporation into protein is mediated via the biological machinery encoded by the ' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Selenocysteine
Selenocysteine (symbol Sec or U, in older publications also as Se-Cys) is the 21st proteinogenic amino acid. Selenoproteins contain selenocysteine residues. Selenocysteine is an analogue of the more common cysteine with selenium in place of the sulfur. Selenocysteine is present in several enzymes (for example glutathione peroxidases, tetraiodothyronine 5′ deiodinases, thioredoxin reductases, formate dehydrogenases, glycine reductases, selenophosphate synthetase 2, methionine-''R''-sulfoxide reductase B1 (SEPX1), and some hydrogenases). It occurs in all three domains of life, including important enzymes (listed above) present in humans. Selenocysteine was discovered by biochemist Thressa Stadtman at the National Institutes of Health. Chemistry Selenocysteine is the Se-analogue of cysteine. It is rarely encountered outside of living tissue (and is not available commercially) because it is very susceptible to air-oxidation. More common is the oxidized derivative selenocystine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Blastocrithidia Nuclear Code
The ''Blastocrithidia'' nuclear code (translation table 31) is a genetic code used by the nuclear genome of the trypanosomatid genus ''Blastocrithidia''. The code (31) : AAs = FFLLSSSSYYEECCWWLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG : Starts = ----------**-----------------------M---------------------------- : Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG : Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG : Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG Bases: adenine (A), cytosine (C), guanine (G) and thymine (T) or uracil (U). Amino acids: Alanine (Ala, A), Arginine (Arg, R), Asparagine (Asn, N), Aspartic acid (Asp, D), Cysteine (Cys, C), Glutamic acid (Glu, E), Glutamine (Gln, Q), Glycine (Gly, G), Histidine (His, H), Isoleucine (Ile, I), Leucine (Leu, L), Lysine (Lys, K), Methionine (Met, M), Phenylalanine (Phe, F), Proline (Pro, P), Serine (Ser, S), T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Condylostoma Nuclear Code
The ''Condylostoma'' nuclear code (translation table 28) is a genetic code used by the nuclear genome of the heterotrich ciliate ''Condylostoma magnum''. The code (28) : AAs = FFLLSSSSYYQQCCWWLLLAPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG : Starts = ----------**--*--------------------M---------------------------- : Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG : Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG : Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG Bases: adenine (A), cytosine (C), guanine (G) and thymine (T) or uracil (U). Amino acids: Alanine (Ala, A), Arginine (Arg, R), Asparagine (Asn, N), Aspartic acid (Asp, D), Cysteine (Cys, C), Glutamic acid (Glu, E), Glutamine (Gln, Q), Glycine (Gly, G), Histidine (His, H), Isoleucine (Ile, I), Leucine (Leu, L), Lysine (Lys, K), Methionine (Met, M), Phenylalanine (Phe, F), Proline (Pro, P), Serine (Ser, S), Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Karyorelict Nuclear Code
The karyorelictid nuclear code (translation table 27) is a genetic code used by the nuclear genome of the Karyorelictea ciliate ''Parduczia'' sp. The code (27) : AAs = FFLLSSSSYYQQCCWWLLLAPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG : Starts = --------------*--------------------M---------------------------- : Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG : Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG : Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG Bases: adenine (A), cytosine (C), guanine (G) and thymine (T) or uracil (U). Amino acids: Alanine (Ala, A), Arginine (Arg, R), Asparagine (Asn, N), Aspartic acid (Asp, D), Cysteine (Cys, C), Glutamic acid (Glu, E), Glutamine (Gln, Q), Glycine (Gly, G), Histidine (His, H), Isoleucine (Ile, I), Leucine (Leu, L), Lysine (Lys, K), Methionine (Met, M), Phenylalanine (Phe, F), Proline (Pro, P), Serine (Ser, S), Threonine ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acids
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling lif ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thraustochytrium Mitochondrial Code
The ''Thraustochytrium'' mitochondrial code (translation table 23) is a genetic code found in the mitochondria of the labyrinthulid protist '' Thraustochytrium aureum''. The mitochondrial genome was sequenced by the Organelle Genome Megasequencing Program. Code Differences from the standard code It is the similar to the bacterial code ( translation table 11) but it contains an additional stop codon (TTA) and also has a different set of start codons. Systematic range and comments * Mitochondria of '' Thraustochytrium aureum''. See also * List of genetic codes While there is much commonality, different parts of the tree of life use slightly different genetic codes. When translating from genome to protein, the use of the correct genetic code is essential. The mitochondrial codes are the relatively well-k ... References External links Organelle Genome Megasequencing Program Molecular genetics Gene expression Protein biosynthesis {{Genetics-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |