|
Somatic Hypermutation
Somatic hypermutation (or SHM) is a cellular mechanism by which the immune system adapts to the new foreign elements that confront it (e.g. microbes), as seen during class switching. A major component of the process of affinity maturation, SHM diversifies B cell receptors used to recognize foreign elements (antigens) and allows the immune system to adapt its response to new threats during the lifetime of an organism. Somatic hypermutation involves a programmed process of mutation affecting the variable regions of immunoglobulin genes. Unlike germline mutation, SHM affects only an organism's individual immune cells, and the mutations are not transmitted to the organism's offspring.Oprea, M. (1999''Antibody Repertoires and Pathogen Recognition:'' The Role of Germline Diversity and Somatic Hypermutation'' (Thesis) University of Leeds.'' Because this mechanism is merely selective and not precisely targeted, somatic hypermutation has been strongly implicated in the development of B-cell l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell (biology)
The cell is the basic structural and functional unit of life forms. Every cell consists of a cytoplasm enclosed within a membrane, and contains many biomolecules such as proteins, DNA and RNA, as well as many small molecules of nutrients and metabolites.Cell Movements and the Shaping of the Vertebrate Body in Chapter 21 of Molecular Biology of the Cell '' fourth edition, edited by Bruce Alberts (2002) published by Garland Science. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos. It is also common to describe small molecules such as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Somatic (biology)
The term somatic - etymologically from the Ancient Greek words of "σωματικός" (sōmatikós, “bodily”) and σῶμα (sôma, “body”) - is often used in biology to refer to the cells of the body in contrast to the reproductive (germline) cells, which usually give rise to the egg or sperm (or other gametes in other organisms). These somatic cells are diploid, containing two copies of each chromosome, whereas germ cells are haploid, as they only contain one copy of each chromosome (in preparation for fertilisation). Although under normal circumstances all somatic cells in an organism contain identical DNA, they develop a variety of tissue-specific characteristics. This process is called differentiation, through epigenetic and regulatory alterations. The grouping of similar cells and tissues creates the foundation for organs. Somatic mutations are changes to the genetics of a multicellular organism that are not passed on to its offspring through the germline. Most canc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Base Excision Repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair pathway repairs bulky helix-distorting lesions. BER is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP sites. These are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch (where a single nucleotide is replaced) or long-patch BER (where 2–10 new nucleotides are synthesized). Lesions processed by BER Single bases in DNA can be chemically damaged by a variety of mechanisms, the most common ones being deamination, oxidation, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Guanine
Guanine () ( symbol G or Gua) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine (uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside is called guanosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with conjugated double bonds. This unsaturated arrangement means the bicyclic molecule is planar. Properties Guanine, along with adenine and cytosine, is present in both DNA and RNA, whereas thymine is usually seen only in DNA, and uracil only in RNA. Guanine has two tautomeric forms, the major keto form (see figures) and rare enol form. It binds to cytosine through three hydrogen bonds. In cytosine, the amino group acts as the hydrogen bond donor and the C-2 carbonyl and the N-3 amine as the hydrogen-bond acceptors. Guanine has the C-6 carbonyl group that acts as the hydrogen bond acceptor, while a group at N ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activation-induced Cytidine Deaminase
Activation-induced cytidine deaminase, also known as AICDA, AID and single-stranded DNA cytosine deaminase, is a 24 kDa enzyme which in humans is encoded by the ''AICDA'' gene. It creates mutations in DNA by deamination of cytosine base, which turns it into uracil (which is recognized as a thymine). In other words, it changes a C:G base pair into a U:G mismatch. The cell's DNA replication machinery recognizes the U as a T, and hence C:G is converted to a T:A base pair. During germinal center development of B lymphocytes, AID also generates other types of mutations, such as C:G to A:T. The mechanism by which these other mutations are created is not well understood. It is a member of the APOBEC family. In B cells in the lymph nodes, AID causes mutations that produce antibody diversity, but that same mutation process leads to B cell lymphoma. Function This gene encodes a DNA-editing deaminase that is a member of the cytidine deaminase family. The protein is involved in somati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Uracil
Uracil () (symbol U or Ura) is one of the four nucleobases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine (T). Uracil is a demethylated form of thymine. Uracil is a common and naturally occurring pyrimidine derivative. The name "uracil" was coined in 1885 by the German chemist Robert Behrend, who was attempting to synthesize derivatives of uric acid. Originally discovered in 1900 by Alberto Ascoli, it was isolated by hydrolysis of yeast nuclein; it was also found in bovine thymus and spleen, herring sperm, and wheat germ. It is a planar, unsaturated compound that has the ability to absorb light. Based on 12C/13C isotopic ratios of organic compounds found in the Murchison meteorite, it is believed that uracil, xanthine, and related molecules can also be formed extraterrestrially. Data from the Cassini mission, orbiting in the Saturn ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytosine
Cytosine () ( symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an amine group at position 4 and a keto group at position 2). The nucleoside of cytosine is cytidine. In Watson-Crick base pairing, it forms three hydrogen bonds with guanine. History Cytosine was discovered and named by Albrecht Kossel and Albert Neumann in 1894 when it was hydrolyzed from calf thymus tissues. A structure was proposed in 1903, and was synthesized (and thus confirmed) in the laboratory in the same year. In 1998, cytosine was used in an early demonstration of quantum information processing when Oxford University researchers implemented the Deutsch-Jozsa algorithm on a two qubit nuclear magnetic resonance quantum computer (NMRQC). In March 2015, NASA scientists reported the formation of cytosine, along with uracil and thym ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deamination
Deamination is the removal of an amino group from a molecule. Enzymes that catalyse this reaction are called deaminases. In the human body, deamination takes place primarily in the liver, however it can also occur in the kidney. In situations of excess protein intake, deamination is used to break down amino acids for energy. The amino group is removed from the amino acid and converted to ammonia. The rest of the amino acid is made up of mostly carbon and hydrogen, and is recycled or oxidized for energy. Ammonia is toxic to the human system, and enzymes convert it to urea or uric acid by addition of carbon dioxide molecules (which is not considered a deamination process) in the urea cycle, which also takes place in the liver. Urea and uric acid can safely diffuse into the blood and then be excreted in urine. Deamination reactions in DNA Cytosine Spontaneous deamination is the hydrolysis reaction of cytosine into uracil, releasing ammonia in the process. This can occur in vitro thr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Uracil
Uracil () (symbol U or Ura) is one of the four nucleobases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine (T). Uracil is a demethylated form of thymine. Uracil is a common and naturally occurring pyrimidine derivative. The name "uracil" was coined in 1885 by the German chemist Robert Behrend, who was attempting to synthesize derivatives of uric acid. Originally discovered in 1900 by Alberto Ascoli, it was isolated by hydrolysis of yeast nuclein; it was also found in bovine thymus and spleen, herring sperm, and wheat germ. It is a planar, unsaturated compound that has the ability to absorb light. Based on 12C/13C isotopic ratios of organic compounds found in the Murchison meteorite, it is believed that uracil, xanthine, and related molecules can also be formed extraterrestrially. Data from the Cassini mission, orbiting in the Saturn ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytosine Chemical Structure
Cytosine () (symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an amine group at position 4 and a keto group at position 2). The nucleoside of cytosine is cytidine. In Watson-Crick base pairing, it forms three hydrogen bonds with guanine. History Cytosine was discovered and named by Albrecht Kossel and Albert Neumann in 1894 when it was hydrolyzed from calf thymus tissues. A structure was proposed in 1903, and was synthesized (and thus confirmed) in the laboratory in the same year. In 1998, cytosine was used in an early demonstration of quantum information processing when Oxford University researchers implemented the Deutsch-Jozsa algorithm on a two qubit nuclear magnetic resonance quantum computer (NMRQC). In March 2015, NASA scientists reported the formation of cytosine, along with uracil and thymin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
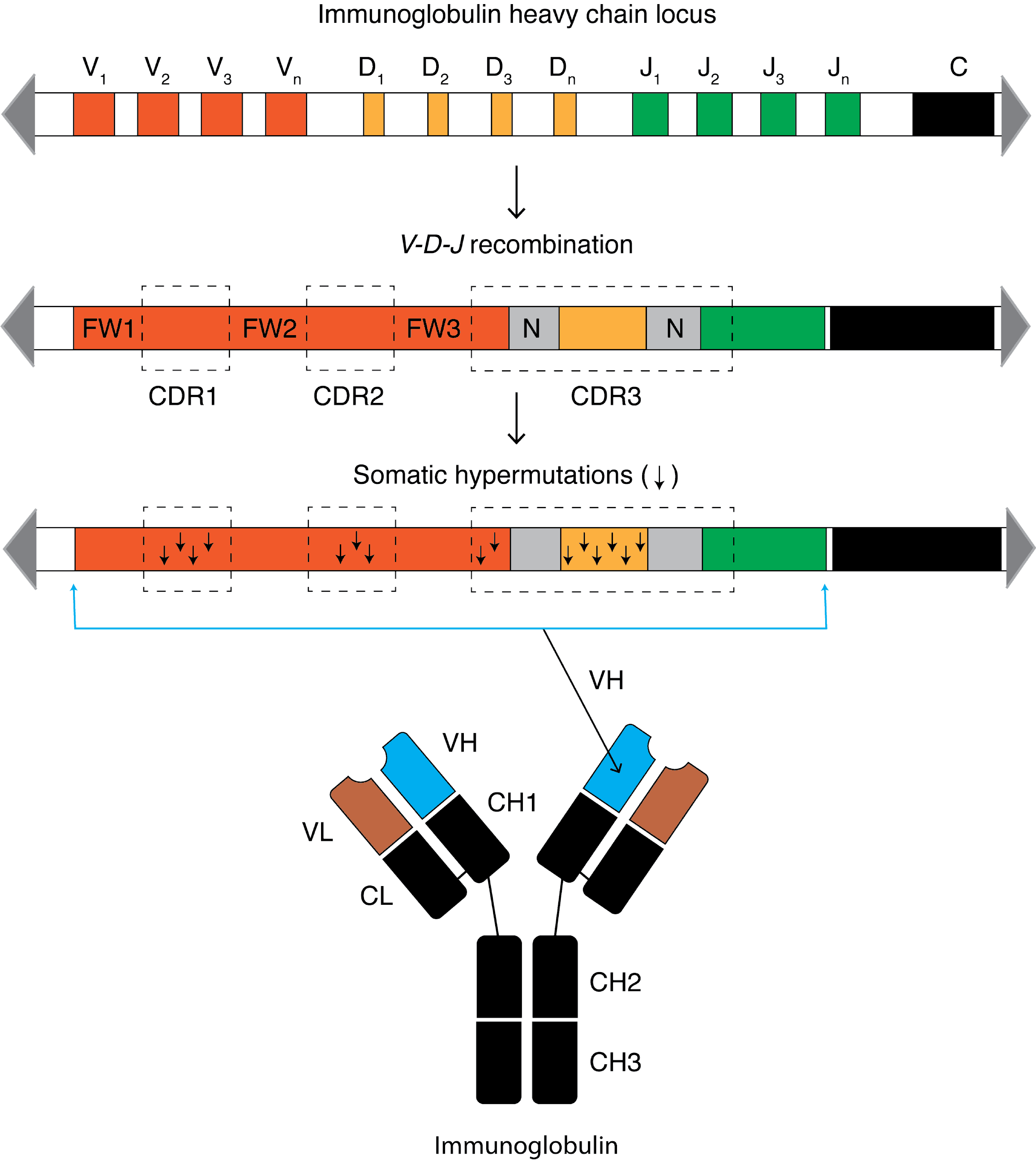
Complementarity-determining Region
Complementarity-determining regions (CDRs) are part of the variable chains in immunoglobulins (antibodies) and T cell receptors, generated by B-cells and T-cells respectively, where these molecules bind to their specific antigen. A set of CDRs constitutes a paratope. As the most variable parts of the molecules, CDRs are crucial to the diversity of antigen specificities generated by lymphocytes. Location and structure There are three CDRs (CDR1, CDR2 and CDR3), arranged non-consecutively, on the amino acid sequence of a variable domain of an antigen receptor. Since the antigen receptors are typically composed of two variable domains (on two different polypeptide chains, heavy and light chain), there are six CDRs for each antigen receptor that can collectively come into contact with the antigen. A single antibody molecule has two antigen receptors and therefore contains twelve CDRs total. There are three CDR loops per variable domain in antibodies. Sixty CDRs can be found on a pen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |