|
Secondary Amino Acids
Secondary amino acids are amino acids which do not contain the amino group but is rather a secondary amine. Secondary amino acids can be classified to cyclic acids such as proline and acyclic N-substituted amino acids. In nature, proline, hydroxyproline, pipecolic acid and sarcosine are well-known secondary amino acids. Proline is the only proteinogenic secondary amino acids. Other secondary amino acids are non-proteinogenic amino acids. In protein, hydroxyproline is incorporated into protein by hydroxylation of proline. Pipecolic acid, a heavier analog of proline, is found in efrapeptin. Sarcosine is a N-methylized glycine so its methyl group is used in many biochemical reactions. Azetidine-2-carboxylic acid, which is a smaller homolog of proline in plants. Properties Proline and its higher homolog pipecolic acid affect the secondary structure of protein. D-alpha-amino acid - L-alpha-amino acid sequence can induce beta hairpin. It suggested that acyclic secondary amino acids a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acids
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling lif ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Amine
In chemistry, amines (, ) are compounds and functional groups that contain a basic nitrogen atom with a lone pair. Amines are formally derivatives of ammonia (), wherein one or more hydrogen atoms have been replaced by a substituent such as an alkyl or aryl group (these may respectively be called alkylamines and arylamines; amines in which both types of substituent are attached to one nitrogen atom may be called alkylarylamines). Important amines include amino acids, biogenic amines, trimethylamine, and aniline; Inorganic derivatives of ammonia are also called amines, such as monochloramine (). The substituent is called an amino group. Compounds with a nitrogen atom attached to a carbonyl group, thus having the structure , are called amides and have different chemical properties from amines. Classification of amines Amines can be classified according to the nature and number of substituents on nitrogen. Aliphatic amines contain only H and alkyl substituents. Aromatic a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Imidic Acid
In chemistry, an imidic acid is any molecule that contains the -C(=NH)-OH functional group. It is the tautomer of an amide and the isomer of an oxime. The term "imino acid" is an obsolete term for this group that should not be used in this context because it has a different molecular structure.{{GoldBookRef , file=I02959 , title=Imino acids , accessdate=2012-04-02 Imidic acids can be formed by metal-catalyzed dehydrogenation of geminal amino alcohols. For example, methanolamine, the parent compound of the amino alcohols, can be dehydrogenated to methanimidic acid, the parent compound of the imidic acids. :H2NCH2OH → HNCHOH + H2 (tautomer of formamide) Geminal amino alcohols with side chains similarly form imidic acids with the same side chains: :H2NCHROH → HNCROH + H2 Another way to form imidic acids is the reaction of carboxylic acids with azanone. For example, the reaction for carbamic acid: :H2NCOOH + HNO → H2NCNHOH + O2 (tautomer of urea) And the general reaction f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Imino Acid
In organic chemistry, an imino acid is any molecule that contains both imine (>C=NH) and carboxyl (-C(=O)-OH) functional groups. Imino acids are structurally related to amino acids, which have amino group instead of imine—a difference of single vs double-bond between nitrogen and carbon. The simplest example is dehydroglycine. D-Amino acid oxidase is an enzyme that is able to convert amino acids into imino acids. Also the direct biosynthetic precursor to the amino acid proline is the imino acid (''S'')-Δ1-pyrroline-5-carboxylate (P5C). Related terminology Secondary amino acids, amino acids containing a secondary amine group are sometimes named imino acids, though this usage is obsolescent. The only proteinogenic amino acid of this type is proline, although the related non-proteinogenic amino acids hydroxyproline and pipecolic acid Pipecolic acid (piperidine-2-carboxylic acid) is an organic compound with the formula HNC5H9CO2H. It is a carboxylic acid derivativ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-methyl-L-amino-acid Oxidase
In enzymology, a N-methyl-L-amino-acid oxidase () is an enzyme that catalyzes the chemical reaction :an N-methyl-L-amino acid + H2O + O2 \rightleftharpoons an L-amino acid + formaldehyde + H2O2 The 3 substrates of this enzyme are N-methyl-L-amino acid, H2O, and O2, whereas its 3 products are L-amino acid, formaldehyde, and H2O2. It has 2 cofactors: FAD, and Flavoprotein. Nomenclature This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-NH group of donors with oxygen as acceptor. The systematic name A systematic name is a name given in a systematic way to one unique group, organism, object or chemical substance, out of a specific population or collection. Systematic names are usually part of a nomenclature. A semisystematic name or semitrivial ... of this enzyme class is N-methyl-L-amino-acid:oxygen oxidoreductase (demethylating). Other names in common use include N-methylamino acid oxidase, and demethylase. References Further ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymology
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ninhydrin
Ninhydrin (2,2-dihydroxyindane-1,3-dione) is an organic compound with the formula C6H4(CO)2C(OH)2. It is used to detect ammonia and amines. Upon reaction with these amines, ninhydrin gets converted into deep blue or purple derivatives, which are called Ruhemann's purple. Ninhydrin is most commonly used to detect fingerprints, as the terminal amines of lysine residues in peptides and proteins sloughed off in fingerprints react with ninhydrin. Ninhydrin is a white solid that is soluble in ethanol and acetone. Ninhydrin can be considered as the hydrate of indane-1,2,3-trione. History Ninhydrin was discovered in 1910 by the German-English chemist Siegfried Ruhemann (1859–1943). In the same year, Ruhemann observed ninhydrin's reaction with amino acids. In 1954, Swedish investigators Oden and von Hofsten proposed that ninhydrin could be used to develop latent fingerprints. Uses Ninhydrin can also be used to monitor deprotection in solid phase peptide synthesis (Kaiser test). The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Efrapeptin C
Efrapeptins are peptides produced by fungi in the genus ''Tolypocladium'' that have antifungal, insecticidal, and mitochondrial ATPase inhibitory activities. They are produced via a biosynthetic pathway similar to but simpler than the Ciclosporin pathway, with nonribosomal peptide synthase (NRPS) and/or polyketide synthase (PKS) being the key elements. The amino acid sequences of efrapeptins are: :Efrapeptin F: Ac-Pip-Aib-Pip-Aib-Aib-Leu-bAla-Gly-Aib-Aib-Pip-Aib-Ala-Leu-Iva-Unk :Efrapeptin G: Ac-Pip-Aib-Pip-Iva-Aib-Leu-bAla-Gly-Aib-Aib-Pip-Aib-Ala-Leu-Iva-Unk ::Aib: 2-methylalanine; Iva: 2-ethylalanine; Unk: does not match to a known amino acid References External links * - Efrapeptin F Efrapeptin Fat ChemSpider ChemSpider is a database of chemicals. ChemSpider is owned by the Royal Society of Chemistry. Database The database contains information on more than 100 million molecules from over 270 data sources including: * EPA DSSTox * U.S. Food and D ... * - Efr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
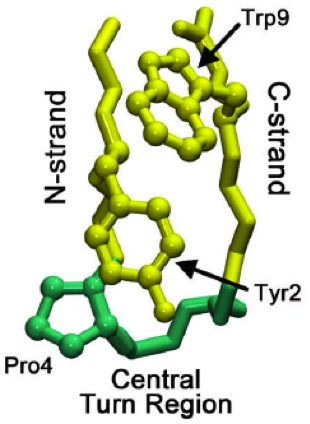
Beta Hairpin
The beta hairpin (sometimes also called beta-ribbon or beta-beta unit) is a simple protein structural motif involving two beta strands that look like a hairpin. The motif consists of two strands that are adjacent in primary structure, oriented in an antiparallel direction (the N-terminus of one sheet is adjacent to the C-terminus of the next), and linked by a short loop of two to five amino acids. Beta hairpins can occur in isolation or as part of a series of hydrogen bonded strands that collectively comprise a beta sheet. Researchers such as Francisco Blanco ''et al.'' have used protein NMR to show that beta-hairpins can be formed from isolated short peptides in aqueous solution, suggesting that hairpins could form nucleation sites for protein folding. Classification Beta hairpins were originally categorized solely by the number of amino acid residues in their loop sequences, such that they were named one-residue, two-residue, etc. This system, however, is somewhat ambiguous ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Azetidine-2-carboxylic Acid
Azetidine-2-carboxylic acid (abbreviated Aze) is a plant Non-proteinogenic amino acids, non-protein amino acid homologue of proline with the molecular formula C4H7NO2. Aze is a heterocyclic, 4 membered ring with nitrogen as its heteroatom (an azetidine), and a carboxylic acid group substituted on one of the ring carbon atoms. The main difference between Aze and proline is the ring of Aze has four members and the ring of proline has five. Aze has the ability to act as an analog of proline and can be incorporated into proteins in place of proline. Synthesis Optically inactive Aze was obtained in small yield from the neurotransmitter GABA by α-bromination, followed by removal of hydrogen bromide from the intermediate γ-amino-α-bromobutyric acid and ring closure by treatment with a barium hydroxide solution. An optically active Aze was obtained by treatment of α,γ-diaminobutyric acid dihydrochloride with a mixture of nitrous and hydrochloric acids to yield γ-amino-α-chlorobu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |