|
ST Motif
The ST motif is a commonly occurring feature in proteins and polypeptides. It consists of four or five amino acid residues with either serine or threonine as the first residue (residue ''i''). It is defined by two internal hydrogen bonds. One is between the side chain oxygen of residue ''i'' and the main chain NH of residue ''i'' + 2 or ''i'' + 3; the other is between the main chain oxygen of residue ''i'' and the main chain NH of residue ''i'' + 3 or ''i'' + 4. Two websites are available for finding and examining ST motifs in proteins, Motivated Proteins: and PDBeMotif. When one of the hydrogen bonds is between the main chain oxygen of residue ''i'' and the side chain NH of residue ''i'' + 3 the motif incorporates a beta turn. When one of the hydrogen bonds is between the side chain oxygen of residue ''i'' and the main chain NH of residue ''i'' + 2 the motif incorporates an ST turn. As with ST turns, a significant propo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residues ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
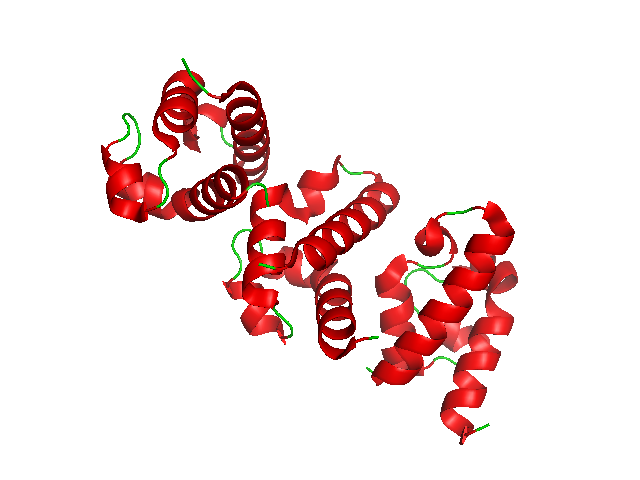
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helicases
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands (hence '' helic- + -ase''), using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases. The human genome codes for 95 non-redundant helicases: 64 RNA helicases and 31 DNA helicases. Many cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair, and ribosome biogenesis involve the separation of nucleic acid strands that necessitates the use of helicases. Some specialized helicases are also involved in sensing of viral nucleic acids during infection and fulfill a immunological function. Function Helicases are oft ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver. Nucleotides are composed of three subunit molecules: a nucleobase, a five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine and thymine; in RNA, uracil is used in place of thymine. Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP) and uridine triphosphate (UTP)—throughout the cell for the many cellular func ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-termini
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that jo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Asparagine
Asparagine (symbol Asn or N) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain carboxamide, classifying it as a polar (at physiological pH), aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it. It is encoded by the codons AAU and AAC. History Asparagine was first isolated in 1806 in a crystalline form by French chemists Louis Nicolas Vauquelin and Pierre Jean Robiquet (then a young assistant). It was isolated from asparagus juice, in which it is abundant, hence the chosen name. It was the first amino acid to be isolated. Three years later, in 1809, Pierre Jean Robiquet identified a substance from liquorice root with properties which he qualified as very similar to those of asparagine, and which Plisson identi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aspartate
Aspartic acid (symbol Asp or D; the ionic form is known as aspartate), is an α-amino acid that is used in the biosynthesis of proteins. Like all other amino acids, it contains an amino group and a carboxylic acid. Its α-amino group is in the protonated –NH form under physiological conditions, while its α-carboxylic acid group is deprotonated −COO− under physiological conditions. Aspartic acid has an acidic side chain (CH2COOH) which reacts with other amino acids, enzymes and proteins in the body. Under physiological conditions (pH 7.4) in proteins the side chain usually occurs as the negatively charged aspartate form, −COO−. It is a non-essential amino acid in humans, meaning the body can synthesize it as needed. It is encoded by the codons GAU and GAC. D-Aspartate is one of two D-amino acids commonly found in mammals. .html" ;"title="/sup>">/sup> In proteins aspartate sidechains are often hydrogen bonded to form asx turns or asx motifs, which frequently occur at t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Asx Motif
The Asx motif is a commonly occurring feature in proteins and polypeptides. It consists of four or five amino acid residues with either aspartate or asparagine as the first residue (residue i). It is defined by two internal hydrogen bonds. One is between the side chain oxygen of residue i and the main chain NH of residue i+2 or i+3; the other is between the main chain oxygen of residue i and the main chain NH of residue i+3 or i+4. Asx motifs occur commonly in proteins and polypeptides. When one of the hydrogen bonds is between the main chain oxygen of residue i and the side chain NH of residue i+3 the motif incorporates a beta turn. When one of the hydrogen bonds is between the side chain oxygen of residue i and the main chain NH of residue i+2 the motif incorporates an Asx turn. As with Asx turns, a significant proportion of Asx motifs occur at the N-terminus of an alpha helix with the Asx as the N cap residue. Asx motifs have thus often been described as helix capping fea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N Cap
The term N cap (N-cap, Ncap) describes an amino acid in a particular position within a protein or polypeptide.{{cite journal, last=Leader, first=DP, author2=Milner-White EJ , title=The structure of the ends of helices in globular proteins, journal=Proteins, year=2011, volume=79, issue=3, pages=1010–1019, doi= 10.1002/prot.22942, pmid=21287629, s2cid=22240314 The N cap residue of an alpha helix is the first amino acid residue at the N terminus of the helix. More precisely, it is defined as the first residue (i) whose CO group is hydrogen-bonded to the NH group of residue i+4 (or sometimes residue i+3). Because of this it is sometimes also described as the residue prior to the helix. Capping motifs are those often found at the N cap. Asx turns, ST turns, and asx motifs are often found at such situations, with the asx or serine or threonine residue at the N cap. The C cap The term C cap (C-cap, Ccap) describes an amino acid in a particular position within a protein or polypeptide.{ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polypeptides
Peptides (, ) are short chains of amino acids linked by peptide bonds. Long chains of amino acids are called proteins. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides. A polypeptide is a longer, continuous, unbranched peptide chain. Hence, peptides fall under the broad chemical classes of biological polymers and oligomers, alongside nucleic acids, oligosaccharides, polysaccharides, and others. A polypeptide that contains more than approximately 50 amino acids is known as a protein. Proteins consist of one or more polypeptides arranged in a biologically functional way, often bound to ligands such as coenzymes and cofactors, or to another protein or other macromolecule such as DNA or RNA, or to complex macromolecular assemblies. Amino acids that have been incorporated into peptides are termed residues. A water molecule is released during formation of each amide bond.. All peptides except cyclic peptides ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ST Turn
The ST turn is a structural feature in proteins and polypeptides. Each consists of three amino acid residues (labeled ''i'', ''i'' + 1 and ''i'' + 2) in which residue ''i'' is a serine (S) or threonine (T) that forms a hydrogen bond from its sidechain oxygen group to the mainchain NH group of residue ''i'' + 2. Similar Structural motif, motifs occur with aspartate or asparagine as residue ''i'', called asx turn. Four types of asx turn and ST turn can be distinguished: types I, I’, II and II’. These categories correspond (via sidechain-mainchain mimicry of residue i) to those of the more abundant hydrogen-bonded beta turns, which have four residues and a hydrogen bond between the CO of residue ''i'' and the NH of residue ''i'' + 3. Regarding their occurrence in proteins, they differ in that type I is the commonest of the four beta turns while type II’ is the commonest of the ST and asx turns. Asx and ST turns both occur frequently at th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |