|
STIP1
Hop, occasionally written HOP, is an abbreviation for Hsp70-Hsp90 Organizing Protein. It functions as a co-chaperone which reversibly links together the protein chaperones Hsp70 and Hsp90. Hop belongs to the large group of co-chaperones, which regulate and assist the major chaperones (mainly heat shock proteins). It is one of the best studied co-chaperones of the Hsp70/Hsp90-complex. It was first discovered in yeast and homologues were identified in human, mouse, rat, insects, plants, parasites, and virus. The family of these proteins is referred to as STI1 (stress inducible protein) and can be divided into yeast, plant, and animal STI1 (Hop). Synonyms Gene The gene for human Hop is located on chromosome 11q13.1 and consists of 14 exons. Structure STI proteins are characterized by some structural features: All homologues have nine tetratricopeptide repeat (TPR) motifs, that are clustered into domains of three TPRs. The TPR motif is a very common structural feature use ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chaperone (protein)
In molecular biology, molecular chaperones are proteins that assist the conformational folding or unfolding of large proteins or macromolecular protein complexes. There are a number of classes of molecular chaperones, all of which function to assist large proteins in proper protein folding during or after synthesis, and after partial denaturation. Chaperones are also involved in the translocation of proteins for proteolysis. The first molecular chaperones discovered were a type of assembly chaperones which assist in the assembly of nucleosomes from folded histones and DNA. One major function of molecular chaperones is to prevent the aggregation of misfolded proteins, thus many chaperone proteins are classified as heat shock proteins, as the tendency for protein aggregation is increased by heat stress. The majority of molecular chaperones do not convey any steric information for protein folding, and instead assist in protein folding by binding to and stabilizing folding interme ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Co-chaperone
Co-chaperones are proteins that assist chaperone (protein), chaperones in protein folding and other functions. Co-chaperones are the non-client binding molecules that assist in protein folding mediated by Hsp70 and Hsp90. They are particularly essential in stimulation of the ATPase activity of these chaperone proteins. There are a great number of different co-chaperones however based on their domain structure most of them fall into two groups: J-domain proteins and tetratricopeptide repeats (TPR). Co-chaperones assist heat shock proteins in the protein folding process. These co-chaperones can function in a number of ways. Primarily co-chaperones are involved in the ATPase functionality of their associated heat shock proteins. Co-chaperones catalyze the hydrolysis ATP to ADP on their respective chaperones which then allows them undergo a large conformational change that allows them to either bind to their substrates with higher affinity or aid in the release of the substrate followin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hsp90
Hsp90 (heat shock protein 90) is a chaperone protein that assists other proteins to fold properly, stabilizes proteins against heat stress, and aids in protein degradation. It also stabilizes a number of proteins required for tumor growth, which is why Hsp90 inhibitors are investigated as anti-cancer drugs. Heat shock proteins, as a class, are among the most highly expressed cellular proteins across all species. As their name implies, heat shock proteins protect cells when stressed by elevated temperatures. They account for 1–2% of total protein in unstressed cells. However, when cells are heated, the fraction of heat shock proteins increases to 4–6% of cellular proteins. Heat shock protein 90 (Hsp90) is one of the most common of the heat-related proteins. The "90" comes from the fact that it has a mass of roughly 90 kilodaltons. A 90 kDa protein is considered fairly large for a non-fibrous protein. Hsp90 is found in bacteria and all branches of eukarya ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hsp70
The 70 kilodalton heat shock proteins (Hsp70s or DnaK) are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms. Intracellularly localized Hsp70s are an important part of the cell's machinery for protein folding, performing chaperoning functions, and helping to protect cells from the adverse effects of physiological stresses. Additionally, membrane-bound Hsp70s have been identified as a potential target for cancer therapies and their extracellularly localized counterparts have been identified as having both membrane-bound and membrane-free structures. Discovery Members of the Hsp70 family are very strongly upregulated by heat stress and toxic chemicals, particularly heavy metals such as arsenic, cadmium, copper, mercury, etc. Heat shock was originally discovered by Ferruccio Ritossa in the 1960s when a lab worker accidentally boosted the incubation temperature of Drosophila (fruit flies). When ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcriptase
A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible. Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host genom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PRNP
Major prion protein (PrP), is encoded in the human by the ''PRNP'' gene also known as CD230 (cluster of differentiation 230). Expression of the protein is most predominant in the nervous system but occurs in many other tissues throughout the body. The protein can exist in multiple isoforms: the normal PrPC form, and the protease-resistant form designated PrPRes such as the disease-causing PrPSc(scrapie) and an isoform located in mitochondria. The misfolded version PrPSc is associated with a variety of cognitive disorders and neurodegenerative diseases such as in animals: ovine scrapie, bovine spongiform encephalopathy (BSE, mad cow disease), feline spongiform encephalopathy, transmissible mink encephalopathy (TME), exotic ungulate encephalopathy, chronic wasting disease (CWD) which affects cervids; and in humans: Creutzfeldt–Jakob disease (CJD), fatal familial insomnia (FFI), Gerstmann–Sträussler–Scheinker syndrome (GSS), kuru, and variant Creutzfeldt–Jakob dis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Piwi-interacting RNA
Piwi-interacting RNA (piRNA) is the largest class of small non-coding RNA molecules expressed in animal cells. piRNAs form RNA-protein complexes through interactions with piwi-subfamily Argonaute proteins. These piRNA complexes are mostly involved in the epigenetic and post-transcriptional silencing of transposable elements and other spurious or repeat-derived transcripts, but can also be involved in the regulation of other genetic elements in germ line cells. piRNAs are mostly created from loci that function as transposon traps which provide a kind of RNA-mediated adaptive immunity against transposon expansions and invasions. They are distinct from microRNA (miRNA) in size (26–31 nucleotides as opposed to 21–24 nt), lack of sequence conservation, increased complexity, and independence of Dicer for biogenesis, at least in animals. (Plant Dcl2 may play a role in rasi/piRNA biogenesis.) Double-stranded RNAs capable of silencing repeat elements, then known as repeat associated s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SiRNA
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA at first non-coding RNA molecules, typically 20-24 (normally 21) base pairs in length, similar to miRNA, and operating within the RNA interference (RNAi) pathway. It interferes with the expression of specific genes with complementary nucleotide sequences by degrading mRNA after transcription, preventing translation. Text was copied from this source, which is available under Creative Commons Attribution 4.0 International License Structure Naturally occurring siRNAs have a well-defined structure that is a short (usually 20 to 24- bp) double-stranded RNA (dsRNA) with phosphorylated 5' ends and hydroxylated 3' ends with two overhanging nucleotides. The Dicer enzyme catalyzes production of siRNAs from long dsRNAs and small hairpin RNAs. siRNAs can also be introduced into cells by transfection. Since in principle any gene can be knocked down by a syntheti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including ''co-suppression'', ''post-transcriptional gene silencing'' (PTGS), and ''quelling''. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm '' Caenorhabditis elegans'', which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
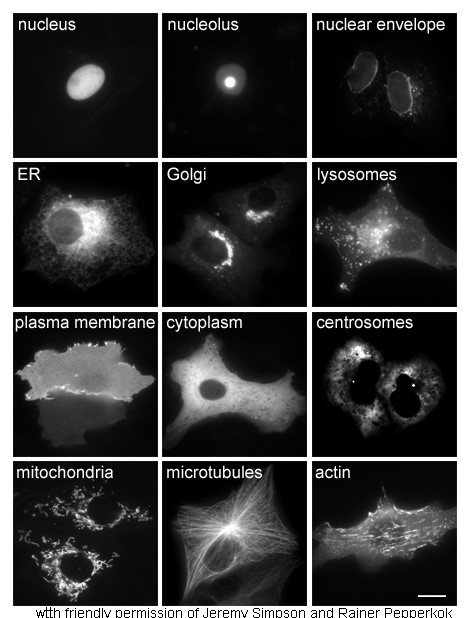
Cell Nucleus
The cell nucleus (pl. nuclei; from Latin or , meaning ''kernel'' or ''seed'') is a membrane-bound organelle found in eukaryotic cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix, a network within the nucleus that adds mechanical support. The cell nucleus contains nearly all of the cell's genome. Nuclear DNA is often organized into multiple chromosomes – long stands of DNA dotted with various proteins, such as histones, that protect and organize the DNA. The genes within these chromosomes are structured in such a way to promote cell function. The nucleus maintains the integrity of genes and controls the activities of the cell by regulating gene expres ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. The main components of the cytoplasm are cytosol (a gel-like substance), the organelles (the cell's internal sub-structures), and various cytoplasmic inclusions. The cytoplasm is about 80% water and is usually colorless. The submicroscopic ground cell substance or cytoplasmic matrix which remains after exclusion of the cell organelles and particles is groundplasm. It is the hyaloplasm of light microscopy, a highly complex, polyphasic system in which all resolvable cytoplasmic elements are suspended, including the larger organelles such as the ribosomes, mitochondria, the plant plastids, lipid droplets, and vacuoles. Most cellular activities take place within the cytoplasm, such as many metabolic pathways including glycolysis, and proces ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |