|
SMAWK Algorithm
The SMAWK algorithm is an algorithm for finding the minimum value in each row of an implicitly-defined totally monotone matrix. It is named after the initials of its five inventors, Peter Shor, Shlomo Moran, Alok Aggarwal, Robert Wilber, and Maria Klawe.. Input For the purposes of this algorithm, a matrix is defined to be monotone if each row's minimum value occurs in a column which is equal to or greater than the column of the previous row's minimum. It is totally monotone if the same property is true for every submatrix (defined by an arbitrary subset of the rows and columns of the given matrix). Equivalently, a matrix is totally monotone if there does not exist a 2×2 submatrix whose row minima are in the top right and bottom left corners. Every Monge array is totally monotone, but not necessarily vice versa. For the SMAWK algorithm, the matrix to be searched should be defined as a function, and this function is given as input to the algorithm (together with the dimensions of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Algorithm
In mathematics and computer science, an algorithm () is a finite sequence of Rigour#Mathematics, mathematically rigorous instructions, typically used to solve a class of specific Computational problem, problems or to perform a computation. Algorithms are used as specifications for performing calculations and data processing. More advanced algorithms can use Conditional (computer programming), conditionals to divert the code execution through various routes (referred to as automated decision-making) and deduce valid inferences (referred to as automated reasoning). In contrast, a Heuristic (computer science), heuristic is an approach to solving problems without well-defined correct or optimal results.David A. Grossman, Ophir Frieder, ''Information Retrieval: Algorithms and Heuristics'', 2nd edition, 2004, For example, although social media recommender systems are commonly called "algorithms", they actually rely on heuristics as there is no truly "correct" recommendation. As an e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
All Nearest Smaller Values
In computer science, the all nearest smaller values problem is the following task: for each position in a sequence of numbers, search among the previous positions for the last position that contains a smaller value. This problem can be solved efficiently both by parallel and non-parallel algorithms: , who first identified the procedure as a useful subroutine for other parallel programs, developed efficient algorithms to solve it in the Parallel Random Access Machine model; it may also be solved in linear time on a non-parallel computer using a stack-based algorithm. Later researchers have studied algorithms to solve it in other models of parallel computation. Example Suppose that the input is the binary van der Corput sequence :0, 8, 4, 12, 2, 10, 6, 14, 1, 9, 5, 13, 3, 11, 7, 15. The first element of the sequence (0) has no previous value. The nearest (only) smaller value previous to 8 and to 4 is 0. All three values previous to 12 are smaller, but the nearest one is 4. Continui ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Thresholding (image Processing)
In digital image processing, thresholding is the simplest method of segmenting images. From a grayscale image, thresholding can be used to create binary images. Definition The simplest thresholding methods replace each pixel in an image with a black pixel if the image intensity I_ is less than a fixed value called the threshold T, or a white pixel if the pixel intensity is greater than that threshold. In the example image on the right, this results in the dark tree becoming completely black, and the bright snow becoming completely white. Automatic thresholding While in some cases, the threshold T can be selected manually by the user, there are many cases where the user wants the threshold to be automatically set by an algorithm. In those cases, the threshold should be the "best" threshold in the sense that the partition of the pixels above and below the threshold should match as closely as possible the actual partition between the two classes of objects represented by those ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Prefix Code
A prefix code is a type of code system distinguished by its possession of the prefix property, which requires that there is no whole Code word (communication), code word in the system that is a prefix (computer science), prefix (initial segment) of any other code word in the system. It is trivially true for fixed-length codes, so only a point of consideration for variable-length code, variable-length codes. For example, a code with code has the prefix property; a code consisting of does not, because "5" is a prefix of "59" and also of "55". A prefix code is a uniquely decodable code: given a complete and accurate sequence, a receiver can identify each word without requiring a special marker between words. However, there are uniquely decodable codes that are not prefix codes; for instance, the reverse of a prefix code is still uniquely decodable (it is a suffix code), but it is not necessarily a prefix code. Prefix codes are also known as prefix-free codes, prefix condition codes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural biology, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix (mathematics), matrix. Gaps are inserted between the Residue (chemistry), residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences such as calculating the Edit distance, distance cost between strings in a natural language, or to display financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence ali ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metabolic reactions, DNA replication, Cell signaling, responding to stimuli, providing Cytoskeleton, structure to cells and Fibrous protein, organisms, and Intracellular transport, transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the Nucleic acid sequence, nucleotide sequence of their genes, and which usually results in protein folding into a specific Protein structure, 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called pep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Nucleic Acid Secondary Structure
Nucleic acid secondary structure is the basepairing interactions within a single nucleic acid polymer or between two polymers. It can be represented as a list of bases which are paired in a nucleic acid molecule. The secondary structures of biological DNAs and RNAs tend to be different: biological DNA mostly exists as fully base paired double helices, while biological RNA is single stranded and often forms complex and intricate base-pairing interactions due to its increased ability to form hydrogen bonds stemming from the extra hydroxyl group in the ribose sugar. In a non-biological context, secondary structure is a vital consideration in the nucleic acid design of nucleic acid structures for DNA nanotechnology and DNA computing, since the pattern of basepairing ultimately determines the overall structure of the molecules. Fundamental concepts Base pairing In molecular biology, two nucleotides on opposite complementary DNA or RNA strands that are connected via hydroge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Word Wrap
Text wrapping, also known as line wrapping, word wrapping or line breaking, is breaking a section of text into lines so that it will fit into the available width of a page, window or other display area. In text display, line wrap is continuing on a new line when a line is full, so that each line fits into the viewable area without overflowing, allowing text to be read from top to bottom without any horizontal scrolling. Word wrap is the additional feature of most text editors, word processors, and web browsers, of breaking lines between words rather than within words, where possible. Word wrap makes it unnecessary to hard-code newline delimiters within paragraphs, and allows the display of text to adapt flexibly and dynamically to displays of varying sizes. Examples Soft and hard returns A soft return or soft wrap is the break resulting from line wrap or word wrap (whether automatic or manual), whereas a hard return or hard wrap is an intentional break, creating a new pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
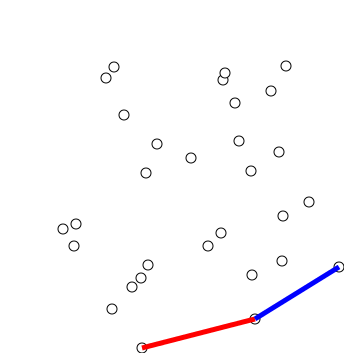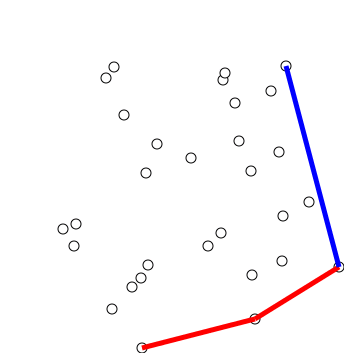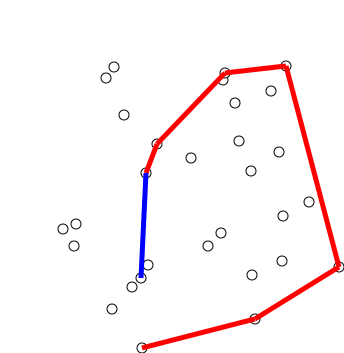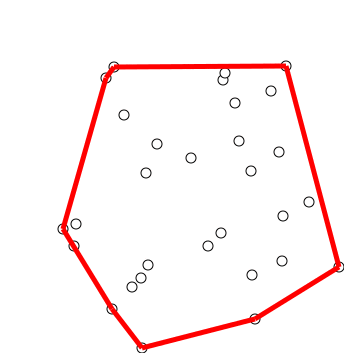
Graham Scan
Graham's scan is a method of finding the convex hull of a finite set of points in the plane with time complexity O(''n'' log ''n''). It is named after Ronald Graham, who published the original algorithm in 1972. The algorithm finds all vertices of the convex hull ordered along its boundary. It uses a stack to detect and remove concavities in the boundary efficiently. Algorithm The first step in this algorithm is to find the point with the lowest y-coordinate. If the lowest y-coordinate exists in more than one point in the set, the point with the lowest x-coordinate out of the candidates should be chosen. Call this point ''P''. This step takes O(''n''), where ''n'' is the number of points in question. Next, the set of points must be sorted in increasing order of the angle they and the point ''P'' make with the x-axis. Any general-purpose sorting algorithm is appropriate for this, for example heapsort (which is O(''n'' log ''n'')). Sorting in order of angle does not require ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Matrix (mathematics)
In mathematics, a matrix (: matrices) is a rectangle, rectangular array or table of numbers, symbol (formal), symbols, or expression (mathematics), expressions, with elements or entries arranged in rows and columns, which is used to represent a mathematical object or property of such an object. For example, \begin1 & 9 & -13 \\20 & 5 & -6 \end is a matrix with two rows and three columns. This is often referred to as a "two-by-three matrix", a " matrix", or a matrix of dimension . Matrices are commonly used in linear algebra, where they represent linear maps. In geometry, matrices are widely used for specifying and representing geometric transformations (for example rotation (mathematics), rotations) and coordinate changes. In numerical analysis, many computational problems are solved by reducing them to a matrix computation, and this often involves computing with matrices of huge dimensions. Matrices are used in most areas of mathematics and scientific fields, either directly ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Stack (data Structure)
In computer science, a stack is an abstract data type that serves as a collection of elements with two main operations: * Push, which adds an element to the collection, and * Pop, which removes the most recently added element. Additionally, a peek operation can, without modifying the stack, return the value of the last element added. The name ''stack'' is an analogy to a set of physical items stacked one atop another, such as a stack of plates. The order in which an element added to or removed from a stack is described as last in, first out, referred to by the acronym LIFO. As with a stack of physical objects, this structure makes it easy to take an item off the top of the stack, but accessing a datum deeper in the stack may require removing multiple other items first. Considered a sequential collection, a stack has one end which is the only position at which the push and pop operations may occur, the ''top'' of the stack, and is fixed at the other end, the ''bottom''. A s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |