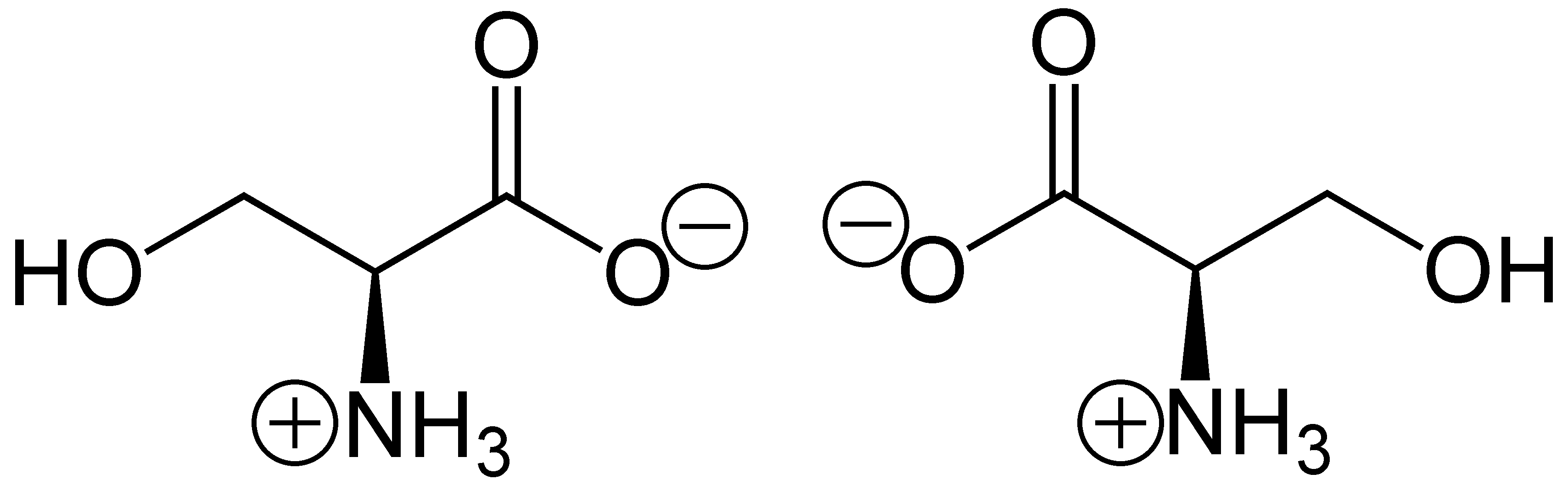|
SHIP1
Src homology 2 (SH2) domain containing inositol polyphosphate 5-phosphatase 1 (SHIP1) is an enzyme with phosphatase activity. SHIP1 is structured by multiple domain and is encoded by the ''INPP5D'' gene in humans. SHIP1 is expressed predominantly by hematopoietic cells but also, for example, by osteoblasts and endothelial cells. This phosphatase is important for the regulation of cellular activation. Not only catalytic but also adaptor activities of this protein are involved in this process. Its movement from the cytosol to the cytoplasmic membrane, where predominantly performs its function, is mediated by tyrosine phosphorylation of the intracellular chains of cell surface receptors that SHIP1 binds. Insufficient regulation of SHIP1 leads to different pathologies. Structure and regulation of activity SHIP1 is a 145 kDa large protein and member of the inositol polyphosphate-5-phosphatase (INPP5) family. Alternate transcriptional splice variants, encoding different isoforms, have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphatidylinositol 3,4-bisphosphate
Phosphatidylinositol (3,4)-bisphosphate (PtdIns(3,4)''P''2) is a minor phospholipid component of cell membranes, yet an important second messenger. The generation of PtdIns(3,4)''P''2 at the plasma membrane activates a number of important cell signaling pathways. Of all the phospholipids found within the membrane, inositol phospholipids make up less than 10%. Phosphoinositide’s (PI’s) also known as phosphatidylinositol phosphates, are synthesized in the cells endoplasmic reticulum by the protein phosphatidylinositol synthase (PIS). PI’s are highly compartmentalized, their main components include a glycerol backbone, two fatty acid chains enriched with stearic acid and arachidonic acid, and an inositol ring whose phosphate groups regulation differs between organelles depending on the specific PI and PIP kinases and PIP phosphatases present in the organelle (Image 1). These kinases and phosphatases conduct phosphorylation and dephosphorylation at the inositol sugar head groups ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphatidylinositol (3,4,5)-trisphosphate
Phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)''P''3), abbreviated PIP3, is the product of the class I phosphoinositide 3-kinases (PI 3-kinases) phosphorylation of phosphatidylinositol (4,5)-bisphosphate (PIP2). It is a phospholipid that resides on the plasma membrane. Discovery In 1988, Lewis C. Cantley published a paper describing the discovery of a novel type of phosphoinositide kinase with the unprecedented ability to phosphorylate the 3' position of the inositol ring resulting in the formation of phosphatidylinositol-3-phosphate (PI3P). Working independently, Alexis Traynor-Kaplan and coworkers published a paper demonstrating that a novel lipid, phosphatidylinositol 3,4,5 trisphosphate (PIP3) occurs naturally in human neutrophils with levels that increased rapidly following physiologic stimulation with chemotactic peptide. Subsequent studies demonstrated that ''in vivo'' the enzyme originally identified by Cantley's group prefers PtdIns(4,5)P2 as a substrate, pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrolysis
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution reaction, substitution, elimination reaction, elimination, and solvation reactions in which water is the nucleophile. Biological hydrolysis is the cleavage of biomolecules where a water molecule is consumed to effect the separation of a larger molecule into component parts. When a carbohydrate is broken into its component sugar molecules by hydrolysis (e.g., sucrose being broken down into glucose and fructose), this is recognized as saccharification. Hydrolysis reactions can be the reverse of a condensation reaction in which two molecules join into a larger one and eject a water molecule. Thus hydrolysis adds water to break down, whereas condensation builds up by removing water. Types Usually hydrolysis is a chemical process in which a molecule of water is added to a substance. Sometimes this addition causes both the substance and w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allosteric Regulation
In biochemistry, allosteric regulation (or allosteric control) is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site. The site to which the effector binds is termed the ''allosteric site'' or ''regulatory site''. Allosteric sites allow effectors to bind to the protein, often resulting in a conformational change and/or a change in protein dynamics. Effectors that enhance the protein's activity are referred to as ''allosteric activators'', whereas those that decrease the protein's activity are called ''allosteric inhibitors''. Allosteric regulations are a natural example of control loops, such as feedback from downstream products or feedforward from upstream substrates. Long-range allostery is especially important in cell signaling. Allosteric regulation is also particularly important in the cell's ability to adjust enzyme activity. The term ''allostery'' comes from the Ancient Greek ''allos'' (), "other", and ''stereos' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Kinase, AMP-activated, Alpha 1
5'-AMP-activated protein kinase catalytic subunit alpha-1 is an enzyme that in humans is encoded by the ''PRKAA1'' gene. The protein encoded by this gene belongs to the serine/threonine protein kinase family. It is the catalytic subunit of the 5'-prime-AMP-activated protein kinase (AMPK). AMPK is a cellular energy sensor conserved in all eukaryotic cells. The kinase activity of AMPK is activated by the stimuli that increase the cellular AMP/ ATP ratio. AMPK regulates the activities of a number of key metabolic enzymes through phosphorylation. It protects cells from stresses that cause ATP depletion by switching off ATP-consuming biosynthetic pathways. Alternatively spliced transcript variants encoding distinct isoforms have been observed. Interactions Protein kinase, AMP-activated, alpha 1 has been shown to interact with TSC2 Tuberous Sclerosis Complex 2 (TSC2), also known as Tuberin, is a protein that in humans is encoded by the ''TSC2'' gene. Function Mutations in this gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L-stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure was estab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphotyrosine-binding Domain
In molecular biology, Phosphotyrosine-binding domains are protein domains which bind to phosphotyrosine. The phosphotyrosine-binding domain (PTB, also phosphotyrosine-interaction or PI domain) in the protein tensin tends to be found at the C-terminus. Tensin is a multi-domain protein that binds to actin filaments and functions as a focal-adhesion molecule (focal adhesions are regions of plasma membrane through which cells attach to the extracellular matrix). Human tensin has actin-binding sites, an SH2 () domain and a region similar to the tumour suppressor PTEN. The PTB domain interacts with the cytoplasmic tails of beta integrin by binding to an NPXY motif. The phosphotyrosine-binding domain of insulin receptor substrate-1 is not related to the phosphotyrosine-binding domain of tensin. Insulin receptor substrate-1 proteins contain both a pleckstrin homology domain and a phosphotyrosine binding (PTB) domain. The PTB domains facilitate interaction with the activated tyrosi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tyrosine
-Tyrosine or tyrosine (symbol Tyr or Y) or 4-hydroxyphenylalanine is one of the 20 standard amino acids that are used by cells to synthesize proteins. It is a non-essential amino acid with a polar side group. The word "tyrosine" is from the Greek ''tyrós'', meaning ''cheese'', as it was first discovered in 1846 by German chemist Justus von Liebig in the protein casein from cheese. It is called tyrosyl when referred to as a functional group or side chain. While tyrosine is generally classified as a Hydrophobe, hydrophobic amino acid, it is more hydrophilic than phenylalanine. It is Genetic code, encoded by the Genetic code#Codons, codons UAC and UAU in messenger RNA. Functions Aside from being a proteinogenic amino acid, tyrosine has a special role by virtue of the phenol functionality. It occurs in proteins that are part of signal transduction processes and functions as a receiver of phosphate groups that are transferred by way of protein kinases. Phosphorylation of the hyd ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SH3 Domain
The SRC Homology 3 Domain (or SH3 domain) is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src. It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase-activating protein, CDC24 and cdc25. SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. The SH3 proteins interact with adaptor proteins and tyrosine kinases. Interacting with tyrosine kinases, SH3 proteins usually bind far away from the active site. Approximately 300 SH3 domains are found in proteins encoded in the human genome. In addition to that, the SH3 domain was responsible for controlling protein-protein interactions in the signal transduction pathways and regulating the interactions ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |