|
SBPAX
BioPAX (Biological Pathway Exchange) is a RDF/OWL-based standard language to represent biological pathways at the molecular and cellular level. Its major use is to facilitate the exchange of pathway data. Pathway data captures our understanding of biological processes, but its rapid growth necessitates development of databases and computational tools to aid interpretation. However, the current fragmentation of pathway information across many databases with incompatible formats presents barriers to its effective use. BioPAX solves this problem by making pathway data substantially easier to collect, index, interpret and share. BioPAX can represent metabolic and signaling pathways, molecular and genetic interactions and gene regulation networks. BioPAX was created through a community process. Through BioPAX, millions of interactions organized into thousands of pathways across many organisms, from a growing number of sources, are available. Thus, large amounts of pathway data are availa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SBPAX
BioPAX (Biological Pathway Exchange) is a RDF/OWL-based standard language to represent biological pathways at the molecular and cellular level. Its major use is to facilitate the exchange of pathway data. Pathway data captures our understanding of biological processes, but its rapid growth necessitates development of databases and computational tools to aid interpretation. However, the current fragmentation of pathway information across many databases with incompatible formats presents barriers to its effective use. BioPAX solves this problem by making pathway data substantially easier to collect, index, interpret and share. BioPAX can represent metabolic and signaling pathways, molecular and genetic interactions and gene regulation networks. BioPAX was created through a community process. Through BioPAX, millions of interactions organized into thousands of pathways across many organisms, from a growing number of sources, are available. Thus, large amounts of pathway data are availa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Systems Biology Ontology
The Systems Biology Ontology (SBO) is a set of controlled, relational vocabularies of terms commonly used in systems biology, and in particular in computational modeling. SBO is part of thBioModels.neteffort. Motivation The rise of Systems Biology, seeking to comprehend biological processes as a whole, highlighted the need to not only develop corresponding quantitative models, but also to create standards allowing their exchange and integration. This concern drove the community to design common data format such as SBML and CellML. SBML is now largely accepted and used in the field. However, as important as the definition of a common syntax is, it is also necessary to make clear the semantics of models. SBO is an attempt to provide the means of annotating models with terms that indicate the intended semantics of an important subset of models in common use in computational systems biology. The development of SBO was first discussed at the 9th SBML Forum Meeting in Heidelberg Oct ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signaling Gateway (website)
Signaling Gateway is a web portal dedicated to signaling pathways powered by the San Diego Supercomputer Center at the University of California, San Diego. It was initiated by a collaboration between the Alliance for Cellular Signaling and Nature. A primary feature is the Molecule Pages database. Molecule Pages Database (online database journal) Signaling Gateway Molecule Pages is a database containing "essential information on more than 8000 mammalian proteins (Mouse and Human) involved in cellular signaling." The content of molecule pages is authored by invited experts and is peer-reviewed. The published pages are citable by digital object identifiers (DOIs). All data in the Molecule Pages are freely available to the public. Data can be exported to PDF, XML, BioPAX/SBPAX and SBML The Systems Biology Markup Language (SBML) is a representation format, based on XML, for communicating and storing computational models of biological processes. It is a free and open standard with ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SABIO-Reaction Kinetics Database
SABIO-RK (System for the Analysis of Biochemical Pathways - Reaction Kinetics) is a web-accessible database storing information about biochemical reactions and their kinetic properties. SABIO-RK comprises a reaction-oriented representation of quantitative information on reaction dynamics based on a given selected publication. This comprises all available kinetic parameters together with their corresponding rate equations, as well as kinetic law and parameter types and experimental and environmental conditions under which the kinetic data were determined. Additionally, SABIO-RK contains information about the underlying biochemical reactions and pathways including their reaction participants, cellular location and detailed information about the enzymes catalysing the reactions. SABIO-RK Database Content The data stored in SABIO-RK in a comprehensive manner is mainly extracted manually from literature. This includes reactions, their participants (substrates, products), modifiers ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Resource Description Framework
The Resource Description Framework (RDF) is a World Wide Web Consortium (W3C) standard originally designed as a data model for metadata. It has come to be used as a general method for description and exchange of graph data. RDF provides a variety of syntax notations and data serialization formats with Turtle (Terse RDF Triple Language) currently being the most widely used notation. RDF is a directed graph composed of triple statements. An RDF graph statement is represented by: 1) a node for the subject, 2) an arc that goes from a subject to an object for the predicate, and 3) a node for the object. Each of the three parts of the statement can be identified by a URI. An object can also be a literal value. This simple, flexible data model has a lot of expressive power to represent complex situations, relationships, and other things of interest, while also being appropriately abstract. RDF was adopted as a W3C recommendation in 1999. The RDF 1.0 specification was published in 2004, th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WikiPathways
WikiPathways is a community resource for contributing and maintaining content dedicated to biological pathways. Any registered WikiPathways user can contribute, and anybody can become a registered user. Contributions are monitored by a group of admins, but the bulk of peer review, editorial curation, and maintenance is the responsibility of the user community. WikiPathways is built using MediaWiki software, a custom graphical pathway editing tool ( PathVisio) and integrated BridgeDb databases covering major gene, protein, and metabolite systems. Pathway content Each article at WikiPathways is dedicated to a particular pathway. Many types of molecular pathways are covered, including metabolic, signaling, regulatory, etc. and the supported species include human, mouse, zebrafish, fruit fly, C. elegans, yeast, rice and arabidopsis, as well as bacteria and plant species. Using a search feature, one can locate a particular pathway by name, by the genes and proteins it contains, or b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Systems Biology
Systems biology is the computational modeling, computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological systems, using a holistic approach (holism instead of the more traditional reductionist, reductionism) to biological research. Particularly from the year 2000 onwards, the concept has been used widely in biology in a variety of contexts. The Human Genome Project is an example of applied systems thinking in biology which has led to new, collaborative ways of working on problems in the biological field of genetics. One of the aims of systems biology is to model and discover emergent property, emergent properties, properties of cell (biology), cells, tissue (biology), tissues and organisms functioning as a system whose theoretical description is only possible using techniques of systems biology. These typically involve metabolic networks or cell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
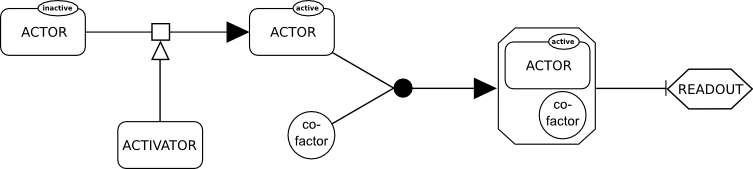
Systems Biology Graphical Notation
The Systems Biology Graphical Notation (SBGN) is a standard graphical representation intended to foster the efficient storage, exchange and reuse of information about signaling pathways, metabolic networks, and gene regulatory networks amongst communities of biochemists, biologists, and theoreticians. The system was created over several years by a community of biochemists, modelers and computer scientists. SBGN is made up of three orthogonal languages for representing different views of biological systems: ''Process Descriptions'', ''Entity Relationships'' and ''Activity Flows''. Each language defines a comprehensive set of symbols with precise semantics, together with detailed syntactic rules regarding the construction and interpretation of maps. Using these three notations, a life scientist can represent in an unambiguous way networks of interactions (for example biochemical interactions). These notations make use of an idea and symbols similar to that used by electrical and ot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MIRIAM
Miriam ( he, מִרְיָם ''Mīryām'', lit. 'Rebellion') is described in the Hebrew Bible as the daughter of Amram and Jochebed, and the older sister of Moses and Aaron. She was a prophetess and first appears in the Book of Exodus. The Torah refers to her as "Miriam the Prophetess" and the Talmud names her as one of the seven major female prophets of Israel. Scripture describes her alongside of Moses and Aaron as delivering the Jews from exile in Egypt: "For I brought you up out of the land of Egypt and redeemed you from the house of slavery, and I sent before you Moses, Aaron, and Miriam". According to the Midrash, just as Moses led the men out of Egypt and taught them Torah, so too Miriam led the women and taught them Torah. Biblical narrative Miriam was the daughter of Amram and Jochebed; she was the sister of Aaron and Moses, the leader of the Israelites in ancient Egypt. The narrative of Moses' infancy in the Torah describes an unnamed sister of Moses observing him ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MIASE
The minimum information about a simulation experiment (MIASE){{cite journal, author1=D. Waltemath , author2=Richard Adams , author3=Daniel A. Beard , author4=rank T. Bergmann , author5=Upinder S. Bhalla , author6=Randall Britten , author7=Vijayalakshmi Chelliah , author8=Michael T. Cooling , author9=Jonathan Cooper , author10=Edmund J. Crampin , author11=Alan Garny , author12=Stefan Hoops , author13=Michael Hucka , author14=Peter Hunter , author15=Edda Klipp , author16=Camille Laibe , author17=Andrew K. Miller , author18=Ion Moraru , author19=David Nickerson , author20=Poul Nielsen , author21=Macha Nikolski , author22=Sven Sahle , author23=Herbert M. Sauro , author24=Henning Schmidt , author25=Jacky L. Snoep , author26=Dominic Tolle , author27=Olaf Wolkenhauer , author28=Nicolas Le Novère , title=Minimum Information About a Simulation Experiment (MIASE). , journal=PLOS Computational Biology , year= 2011 , volume= 7 , issue= 4 , doi=10.1371/journal.pcbi.1001122 , pmid=21552546 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CellML
CellML is an XML based markup language for describing mathematical models. Although it could theoretically describe any mathematical model, it was originally created with the Physiome Project in mind, and hence used primarily to describe models relevant to the field of biology. This is reflected in its name CellML, although this is simply a name, not an abbreviation. CellML is growing in popularity as a portable description format for computational models, and groups throughout the world are using CellML for modelling or developing software tools based on CellML. CellML is similar to Systems Biology Markup Language SBML but provides greater scope for model modularity and reuse, and is not specific to descriptions of biochemistry. History The CellML language grew from a need to share models of cardiac cell dynamics among researchers at a number of sites across the world. The original working group formed in 1998 consisted of David Bullivant, Warren Hedley, and Poul Nielsen; all th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BioModels Database
BioModels is a free and open-source repository for storing, exchanging and retrieving quantitative models of biological interest created in 2006. All the models in the curated section of BioModels Database have been described in peer-reviewed scientific literature. The models stored in BioModels' curated branch are compliant with MIRIAM, the standard of model curation and annotation. The models have been simulated by curators to check that when run in simulations, they provide the same results as described in the publication. Model components are annotated, so the users can conveniently identify each model element and retrieve further information from other resources. Modellers can submit the models in SBML and CellML. Models can subsequently be downloaded in SBMLVCML |