|
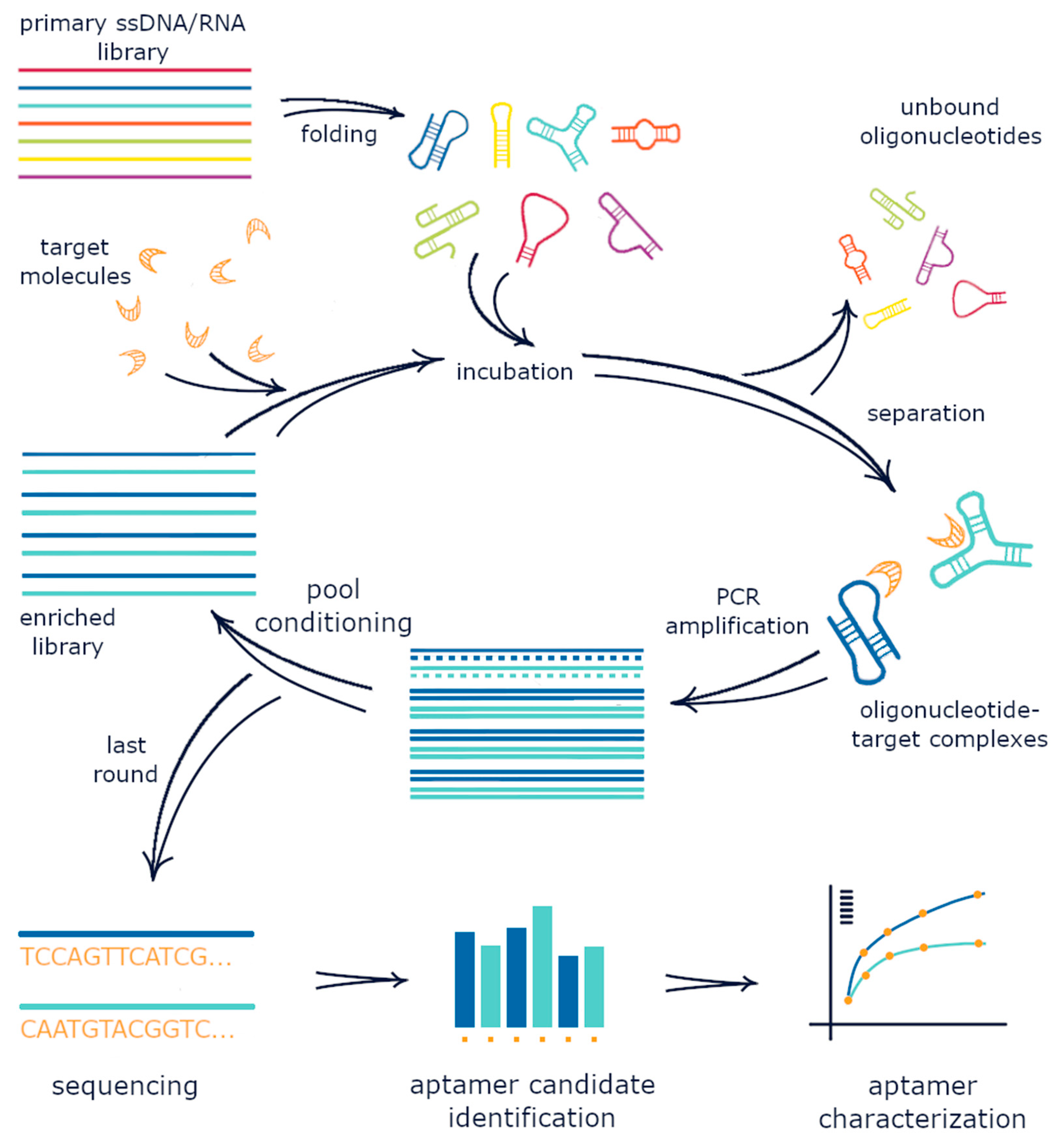
Systematic Evolution Of Ligands By Exponential Enrichment
Systematic evolution of ligands by exponential enrichment (SELEX), also referred to as '' in vitro selection'' or '' in vitro evolution'', is a combinatorial chemistry technique in molecular biology for producing oligonucleotides of either single-stranded DNA or RNA that specifically bind to a target ligand or ligands. These single-stranded DNA or RNA are commonly referred to as aptamers. Although SELEX has emerged as the most commonly used name for the procedure, some researchers have referred to it as SAAB (selected and amplified binding site) and CASTing (cyclic amplification and selection of targets) SELEX was first introduced in 1990. In 2015, a special issue was published in the Journal of Molecular Evolution in the honor of quarter century of the discovery of SELEX. The process begins with the synthesis of a very large oligonucleotide library, consisting of randomly generated sequences of fixed length flanked by constant 5' and 3' ends. The constant ends serve as prim ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SELEX Schematic
SELEX may refer to one of the following: * Selex ES, previously Finmeccanica's defence and security electronics business, now merged into Leonardo-Finmeccanica (new name of Finmeccanica) * SELEX Sistemi Integrati, previously Finmeccanica's civil and military radar company, which became part of Selex ES (in turn merged into Leonardo-Finmeccanica) * SELEX Elsag, previously Finmeccanica's military and civil communications systems supplier, which became part of Selex ES (in turn merged into Leonardo-Finmeccanica) * Systematic evolution of ligands by exponential enrichment, a molecular biology technique used in drug development * Fermilab#Experiments, SEgmented Large-X baryon spectrometer EXperiment, a Fermilab study * Selex Competition, a former constructor of racing cars {{disambig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serum (blood)
Serum () is the fluid and solute component of blood which does not play a role in Coagulation, clotting. It may be defined as blood plasma without the clotting factors, or as blood with all cells and clotting factors removed. Serum includes all proteins not used in Coagulation, blood clotting; all electrolytes, antibodies, antigens, hormones; and any exogenous substances (e.g., drugs or microorganisms). Serum does not contain white blood cells (leukocytes), red blood cells (erythrocytes), platelets, or clotting factors. The study of serum is serology. Serum is used in numerous diagnostic tests as well as blood typing. Measuring the concentration of various molecules can be useful for many applications, such as determining the therapeutic index of a drug candidate in a clinical trial. To obtain serum, a blood sample is allowed to clot (coagulation). The sample is then centrifuged to remove the clot and blood cells, and the resulting liquid wikt:supernatant, supernatant is serum. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Triphosphate
Adenosine triphosphate (ATP) is an organic compound that provides energy to drive many processes in living cells, such as muscle contraction, nerve impulse propagation, condensate dissolution, and chemical synthesis. Found in all known forms of life, ATP is often referred to as the "molecular unit of currency" of intracellular energy transfer. When consumed in metabolic processes, it converts either to adenosine diphosphate (ADP) or to adenosine monophosphate (AMP). Other processes regenerate ATP. The human body recycles its own body weight equivalent in ATP each day. It is also a precursor to DNA and RNA, and is used as a coenzyme. From the perspective of biochemistry, ATP is classified as a nucleoside triphosphate, which indicates that it consists of three components: a nitrogenous base (adenine), the sugar ribose, and the Polyphosphate, triphosphate. Structure ATP consists of an adenine attached by the 9-nitrogen atom to the 1′ carbon atom of a sugar (ribose), which i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Small Molecule
Within the fields of molecular biology and pharmacology, a small molecule or micromolecule is a low molecular weight (≤ 1000 daltons) organic compound that may regulate a biological process, with a size on the order of 1 nm. Many drugs are small molecules; the terms are equivalent in the literature. Larger structures such as nucleic acids and proteins, and many polysaccharides are not small molecules, although their constituent monomers (ribo- or deoxyribonucleotides, amino acids, and monosaccharides, respectively) are often considered small molecules. Small molecules may be used as research tools to probe biological function as well as leads in the development of new therapeutic agents. Some can inhibit a specific function of a protein or disrupt protein–protein interactions. Pharmacology usually restricts the term "small molecule" to molecules that bind specific biological macromolecules and act as an effector, altering the activity or function of the target. Small ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epitope
An epitope, also known as antigenic determinant, is the part of an antigen that is recognized by the immune system, specifically by antibodies, B cells, or T cells. The epitope is the specific piece of the antigen to which an antibody binds. The part of an antibody that binds to the epitope is called a paratope. Although epitopes are usually non-self proteins, sequences derived from the host that can be recognized (as in the case of autoimmune diseases) are also epitopes. The epitopes of protein antigens are divided into two categories, conformational epitopes and linear epitopes, based on their structure and interaction with the paratope. Conformational and linear epitopes interact with the paratope based on the 3-D conformation adopted by the epitope, which is determined by the surface features of the involved epitope residues and the shape or tertiary structure of other segments of the antigen. A conformational epitope is formed by the 3-D conformation adopted by the interac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exodeoxyribonuclease
Exodeoxyribonucleases are both exonucleases and deoxyribonucleases. They catalyze digestion of the ends of linear DNA. They are a type of esterase. They are classified EC 3.1.11. See also * Deoxyribonuclease Deoxyribonuclease (DNase, for short) refers to a group of glycoprotein endonucleases which are enzymes that catalyze the hydrolytic cleavage of phosphodiester linkages in the DNA backbone, thus degrading DNA. The role of the DNase enzyme in cells ... External links * EC 3.1 Deoxyribonucleases {{3.1-enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcriptase
A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible. Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host genom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Binding Affinity
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure. Binding occurs by intermolecular forces, such as ionic bonds, hydrogen bonds and Van der Waals forces. The association or docking is actually reversible through dissociation. Measurably irreversible covalent bonding between a ligand and target molecule is atypical in biological systems. In contrast to the definition of ligan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electric-field Screening
In physics, screening is the damping of electric fields caused by the presence of mobile charge carriers. It is an important part of the behavior of charge-carrying fluids, such as ionized gases (classical plasmas), electrolytes, and charge carriers in electronic conductors (semiconductors, metals). In a fluid, with a given permittivity , composed of electrically charged constituent particles, each pair of particles (with charges and ) interact through the Coulomb force as \mathbf = \frac\hat, where the vector is the relative position between the charges. This interaction complicates the theoretical treatment of the fluid. For example, a naive quantum mechanical calculation of the ground-state energy density yields infinity, which is unreasonable. The difficulty lies in the fact that even though the Coulomb force diminishes with distance as , the average number of particles at each distance is proportional to , assuming the fluid is fairly isotropic. As a result, a charge fl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Confluency
In cell culture biology, confluence refers to the percentage of the surface of a culture dish that is covered by adherent cells. For example, 50 percent confluence means roughly half of the surface is covered, while 100 percent confluence means the surface is completely covered by the cells, and no more room is left for the cells to grow as a monolayer. The cell number refers to, trivially, the number of cells in a given region. Impact on research Many cell lines exhibit differences in growth rate or gene expression depending on the degree of confluence. Cells are typically passaged before becoming fully confluent in order to maintain their proliferation phenotype. Some cell types are not limited by contact inhibition, such as immortalized cells, and may continue to divide and form layers on top of the parent cells. To achieve optimal and consistent results, experiments are usually performed using cells at a particular confluence, depending on the cell type. Extracellular expo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |