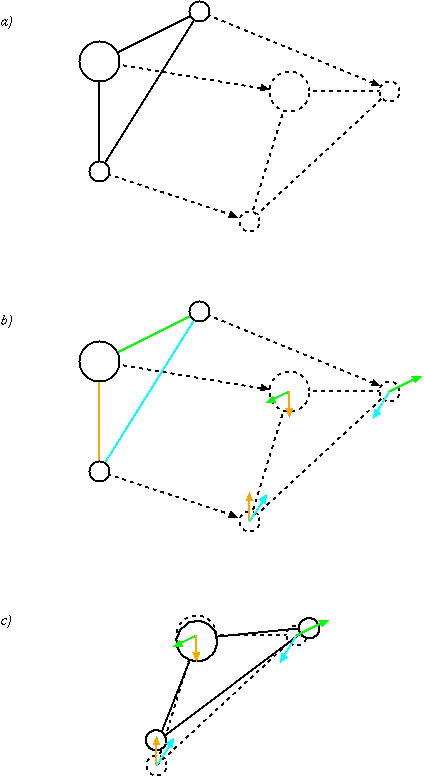
|
SHAKE Algorithm
In computational chemistry, a constraint algorithm is a method for satisfying the Newtonian motion of a rigid body which consists of mass points. A restraint algorithm is used to ensure that the distance between mass points is maintained. The general steps involved are: (i) choose novel unconstrained coordinates (internal coordinates), (ii) introduce explicit constraint forces, (iii) minimize constraint forces implicitly by the technique of Lagrange multipliers or projection methods. Constraint algorithms are often applied to molecular dynamics simulations. Although such simulations are sometimes performed using internal coordinates that automatically satisfy the bond-length, bond-angle and torsion-angle constraints, simulations may also be performed using explicit or implicit constraint forces for these three constraints. However, explicit constraint forces give rise to inefficiency; more computational power is required to get a trajectory of a given length. Therefore, internal co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Computational Chemistry
Computational chemistry is a branch of chemistry that uses computer simulation to assist in solving chemical problems. It uses methods of theoretical chemistry, incorporated into computer programs, to calculate the structures and properties of molecules, groups of molecules, and solids. It is essential because, apart from relatively recent results concerning the hydrogen molecular ion ( dihydrogen cation, see references therein for more details), the quantum many-body problem cannot be solved analytically, much less in closed form. While computational results normally complement the information obtained by chemical experiments, it can in some cases predict hitherto unobserved chemical phenomena. It is widely used in the design of new drugs and materials. Examples of such properties are structure (i.e., the expected positions of the constituent atoms), absolute and relative (interaction) energies, electronic charge density distributions, dipoles and higher multipole moments ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biphenyl
Biphenyl (also known as diphenyl, phenylbenzene, 1,1′-biphenyl, lemonene or BP) is an organic compound that forms colorless crystals. Particularly in older literature, compounds containing the functional group consisting of biphenyl less one hydrogen (the site at which it is attached) may use the prefixes xenyl or diphenylyl. It has a distinctively pleasant smell. Biphenyl is an aromatic hydrocarbon with a molecular formula (C6H5)2. It is notable as a starting material for the production of polychlorinated biphenyls (PCBs), which were once widely used as dielectric fluids and heat transfer agents. Biphenyl is also an intermediate for the production of a host of other organic compounds such as emulsifiers, optical brighteners, crop protection products, and plastics. Biphenyl is insoluble in water, but soluble in typical organic solvents. The biphenyl molecule consists of two connected phenyl rings. Properties and occurrence Biphenyl occurs naturally in coal ta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics. Because molecular systems typically consist of a vast number of particles, it is impossible to determine the properties of such complex systems analytically; MD simulation circumvents this problem by using numerical methods. However, long MD simulations are mathematically ill-conditioned, generating cumulative err ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mechanics
Mechanics (Greek: ) is the area of mathematics and physics concerned with the relationships between force, matter, and motion among physical objects. Forces applied to objects result in displacements, or changes of an object's position relative to its environment. Theoretical expositions of this branch of physics has its origins in Ancient Greece, for instance, in the writings of Aristotle and Archimedes (see History of classical mechanics and Timeline of classical mechanics). During the early modern period, scientists such as Galileo, Kepler, Huygens, and Newton laid the foundation for what is now known as classical mechanics. As a branch of classical physics, mechanics deals with bodies that are either at rest or are moving with velocities significantly less than the speed of light. It can also be defined as the physical science that deals with the motion of and forces on bodies not in the quantum realm. History Antiquity The ancient Greek philosophers were among ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eigenvalues
In linear algebra, an eigenvector () or characteristic vector of a linear transformation is a nonzero vector that changes at most by a scalar factor when that linear transformation is applied to it. The corresponding eigenvalue, often denoted by \lambda, is the factor by which the eigenvector is scaled. Geometrically, an eigenvector, corresponding to a real nonzero eigenvalue, points in a direction in which it is stretched by the transformation and the eigenvalue is the factor by which it is stretched. If the eigenvalue is negative, the direction is reversed. Loosely speaking, in a multidimensional vector space, the eigenvector is not rotated. Formal definition If is a linear transformation from a vector space over a field into itself and is a nonzero vector in , then is an eigenvector of if is a scalar multiple of . This can be written as T(\mathbf) = \lambda \mathbf, where is a scalar in , known as the eigenvalue, characteristic value, or characteristic root ass ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Linear Convergence
In numerical analysis, the order of convergence and the rate of convergence of a convergent sequence are quantities that represent how quickly the sequence approaches its limit. A sequence (x_n) that converges to x^* is said to have ''order of convergence'' q \geq 1 and ''rate of convergence'' \mu if : \lim _ \frac=\mu. The rate of convergence \mu is also called the ''asymptotic error constant''. Note that this terminology is not standardized and some authors will use ''rate'' where this article uses ''order'' (e.g., ). In practice, the rate and order of convergence provide useful insights when using iterative methods for calculating numerical approximations. If the order of convergence is higher, then typically fewer iterations are necessary to yield a useful approximation. Strictly speaking, however, the asymptotic behavior of a sequence does not give conclusive information about any finite part of the sequence. Similar concepts are used for discretization methods. The solutio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Giovanni Ciccotti
Giovanni Ciccotti (born 19 December 1943, in Rome Italy) is an Italian physicist. Ciccotti held the position of Professor of the Structure of Matter at the University of Rome La Sapienza until 2013. He is the author of more than a hundred articles on molecular dynamics and statistical mechanics. He worked with J.P. Ryckaert JP may refer to: Arts and media * ''JP'' (album), 2001, by American singer Jesse Powell * ''Jp'' (magazine), an American Jeep magazine * '' Jönköpings-Posten'', a Swedish newspaper * Judas Priest, an English heavy metal band * ''Jurassic Par ... on new methods for molecular dynamics on constrained systems. See also SHAKE algorithm. He has edited several books on molecular dynamics and statistical mechanics developments, including: # "Molecular Dynamics Simulation of Statistical Mechanical Systems", E. Fermi 1985 Summer School. G. Ciccotti and W. G. Hoover Eds. North Holland, Amsterdam, 1986. # "Simulation of Liquids and Solids. Molecular Dynamics a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quadratic Convergence
In numerical analysis, the order of convergence and the rate of convergence of a convergent sequence are quantities that represent how quickly the sequence approaches its limit. A sequence (x_n) that converges to x^* is said to have ''order of convergence'' q \geq 1 and ''rate of convergence'' \mu if : \lim _ \frac=\mu. The rate of convergence \mu is also called the ''asymptotic error constant''. Note that this terminology is not standardized and some authors will use ''rate'' where this article uses ''order'' (e.g., ). In practice, the rate and order of convergence provide useful insights when using iterative methods for calculating numerical approximations. If the order of convergence is higher, then typically fewer iterations are necessary to yield a useful approximation. Strictly speaking, however, the asymptotic behavior of a sequence does not give conclusive information about any finite part of the sequence. Similar concepts are used for discretization methods. The soluti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LU Decomposition
In numerical analysis and linear algebra, lower–upper (LU) decomposition or factorization factors a matrix as the product of a lower triangular matrix and an upper triangular matrix (see matrix decomposition). The product sometimes includes a permutation matrix as well. LU decomposition can be viewed as the matrix form of Gaussian elimination. Computers usually solve square systems of linear equations using LU decomposition, and it is also a key step when inverting a matrix or computing the determinant of a matrix. The LU decomposition was introduced by the Polish mathematician Tadeusz Banachiewicz in 1938. Definitions Let ''A'' be a square matrix. An LU factorization refers to the factorization of ''A'', with proper row and/or column orderings or permutations, into two factors – a lower triangular matrix ''L'' and an upper triangular matrix ''U'': : A = LU. In the lower triangular matrix all elements above the diagonal are zero, in the upper triangular matrix, all ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecule
A molecule is a group of two or more atoms held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions which satisfy this criterion. In quantum physics, organic chemistry, and biochemistry, the distinction from ions is dropped and ''molecule'' is often used when referring to polyatomic ions. A molecule may be homonuclear, that is, it consists of atoms of one chemical element, e.g. two atoms in the oxygen molecule (O2); or it may be heteronuclear, a chemical compound composed of more than one element, e.g. water (molecule), water (two hydrogen atoms and one oxygen atom; H2O). In the kinetic theory of gases, the term ''molecule'' is often used for any gaseous particle regardless of its composition. This relaxes the requirement that a molecule contains two or more atoms, since the noble gases are individual atoms. Atoms and complexes connected by non-covalent interactions, such as hydrogen bonds or ionic bonds, are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Velocity Verlet
Verlet integration () is a numerical method used to integrate Newton's equations of motion. It is frequently used to calculate trajectories of particles in molecular dynamics simulations and computer graphics. The algorithm was first used in 1791 by Jean Baptiste Delambre and has been rediscovered many times since then, most recently by Loup Verlet in the 1960s for use in molecular dynamics. It was also used by P. H. Cowell and A. C. C. Crommelin in 1909 to compute the orbit of Halley's Comet, and by Carl Størmer in 1907 to study the trajectories of electrical particles in a magnetic field (hence it is also called Störmer's method). The Verlet integrator provides good numerical stability, as well as other properties that are important in physical systems such as time reversibility and preservation of the symplectic form on phase space, at no significant additional computational cost over the simple Euler method. Basic Störmer–Verlet For a second-order differential equa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid resid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |