|
Rapid Deployment Vaccine Collaborative
The Rapid Deployment Vaccine Collaborative (RaDVaC), is a non-profit, collaborative, open-source vaccine research organization founded in March 2020 by Preston Estep and colleagues from various fields of expertise, motivated to respond to the COVID-19 pandemic through rapid, adaptable, transparent, and accessible vaccine development. The members of RaDVaC contend that even the accelerated vaccine approvals, such as the FDA's Emergency Use Authorization, does not make vaccines available quickly enough. The core group has published a series of white papers online, detailing both the technical principles of and protocols for their research vaccine formulas, as well as dedicated materials and protocols pages. All of the organization's published work has been released under Creative Commons non-commercial licenses, including those contributing to the Open COVID Pledge. Multiple individuals involved with the project have engaged in self-experimentation to assess vaccine safety and effica ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RaDVaC Logo (Small)
The Rapid Deployment Vaccine Collaborative (RaDVaC), is a non-profit, collaborative, open-source vaccine research organization founded in March 2020 by Preston Estep and colleagues from various fields of expertise, motivated to respond to the COVID-19 pandemic through rapid, adaptable, transparent, and accessible vaccine development. The members of RaDVaC contend that even the accelerated vaccine approvals, such as the FDA's Emergency Use Authorization, does not make vaccines available quickly enough. The core group has published a series of white papers online, detailing both the technical principles of and protocols for their research vaccine formulas, as well as dedicated materials and protocols pages. All of the organization's published work has been released under Creative Commons non-commercial licenses, including those contributing to the Open COVID Pledge. Multiple individuals involved with the project have engaged in self-experimentation to assess vaccine safety and effica ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
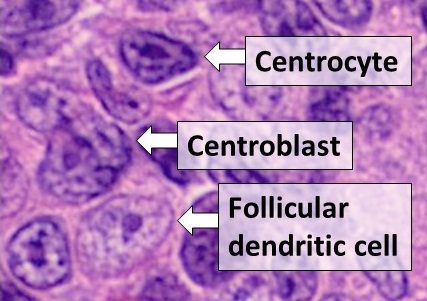
Dendritic Cell
Dendritic cells (DCs) are antigen-presenting cells (also known as ''accessory cells'') of the mammalian immune system. Their main function is to process antigen material and present it on the cell surface to the T cells of the immune system. They act as messengers between the innate and the adaptive immune systems. Dendritic cells are present in those tissues that are in contact with the external environment, such as the skin (where there is a specialized dendritic cell type called the Langerhans cell) and the inner lining of the nose, lungs, stomach and intestines. They can also be found in an immature state in the blood. Once activated, they migrate to the lymph nodes where they interact with T cells and B cells to initiate and shape the adaptive immune response. At certain development stages they grow branched projections, the ''dendrites'' that give the cell its name (δένδρον or déndron being Greek for 'tree'). While similar in appearance, these are structures ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
COVID-19 Vaccine Producers
Coronavirus disease 2019 (COVID-19) is a contagious disease caused by a virus, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The first known case was identified in Wuhan, China, in December 2019. The disease quickly spread worldwide, resulting in the COVID-19 pandemic. The symptoms of COVID‑19 are variable but often include fever, cough, headache, fatigue, breathing difficulties, loss of smell, and loss of taste. Symptoms may begin one to fourteen days after exposure to the virus. At least a third of people who are infected do not develop noticeable symptoms. Of those who develop symptoms noticeable enough to be classified as patients, most (81%) develop mild to moderate symptoms (up to mild pneumonia), while 14% develop severe symptoms (dyspnea, hypoxia, or more than 50% lung involvement on imaging), and 5% develop critical symptoms (respiratory failure, shock, or multiorgan dysfunction). Older people are at a higher risk of developing severe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Vitalik Buterin
Vitaly "Vitalik" Buterin (born 1994) is a Russian-Canadian computer programmer and founder of Ethereum. Buterin became involved with cryptocurrency early in its inception, co-founding ''Bitcoin Magazine'' in 2011. In 2014, Buterin deployed Ethereum on blockchain with Dimitry Buterin, Gavin Wood, Charles Hoskinson, Anthony Di Iorio and Joseph Lubin (entrepreneur), Joseph Lubin. Early life and education Buterin was born in Kolomna, Russia, in 1994. His father was a computer scientist. He lived in the area until the age of six when his parents Immigration to Canada, emigrated to Canada in search of better employment opportunities. While in grade three of elementary school in Canada, Buterin was placed into a class for gifted children and was drawn to mathematics, programming, and economics. Buterin then attended The Abelard School, a private high school in Toronto. Buterin learned about Bitcoin, from his father, Dimitry Buterin at the age of 17. After high school, Buterin attend ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Challenge Study
A human challenge study, also called a challenge trial or controlled human infection model (CHIM), is a type of clinical trial for a vaccine or other pharmaceutical involving the intentional exposure of the test subject to the condition tested. Human challenge studies may be ethically controversial because they involve exposing test subjects to dangers beyond those posed by potential side effects of the substance being tested. During the mid 20th and 21st century, the number of human challenge studies has been increasing. A challenge study to test promising vaccines for prevention of COVID-19 was under consideration during 2020 by several vaccine developers, including the World Health Organization (WHO), and was approved in the UK in 2021. Over the second half of the 20th and the 21st centuries, vaccines for some 15 major pathogens have been fast-tracked in human challenge studies – involving about 30,000 participants – while contributing toward vaccine development to prevent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SARS-CoV-2 Omicron Variant
Omicron (B.1.1.529) is a variant of SARS-CoV-2 first reported to the World Health Organization (WHO) by the Network for Genomics Surveillance in South Africa on 24 November 2021. It was first detected in Botswana and has spread to become the predominant variant in circulation around the world. Following the original BA.1 variant, several subvariants of Omicron have emerged: BA.2, BA.3, BA.4, and BA.5. Since October 2022, two subvariants of BA.5 named BQ.1 and BQ.1.1 have spread rapidly. Three doses of a COVID-19 vaccine provide protection against severe disease and hospitalisation caused by BA.1 and BA.2. The immunity effects of BA.2 are similar to those of BA.1. For three-dose vaccinated individuals, the BA.4 and BA.5 variants are more infectious than previous subvariants, making it likely, , for a new peak in COVID-19 infections to occur. __TOC__ Classification On 26 November, the WHO's Technical Advisory Group on SARS-CoV-2 Virus Evolution declared PANGO linea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SARS-CoV-2 Mu Variant
The Mu variant, also known as lineage B.1.621 or VUI-21JUL-1, is one of the variants of SARS-CoV-2, the virus that causes COVID-19. It was first detected in Colombia in January 2021 and was designated by the WHO as a variant of interest on August 30, 2021. The WHO said the variant has mutations that indicate a risk of resistance to the current vaccines and stressed that further studies were needed to better understand it. Outbreaks of the Mu variant were reported in South America and Europe. The B.1.621 lineage has a sublineage, labeled B.1.621.1 under the PANGO nomenclature, which has already been detected in more than 20 countries worldwide. Under the simplified naming scheme proposed by the World Health Organization, B.1.621 was labeled "Mu variant", and was considered a variant of interest (VOI), but not a variant of concern. Classification Naming In January 2021, the lineage was first documented in Colombia and was named as ''lineage B.1.621''. On July 1, 2021, Public ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SARS-CoV-2 Gamma Variant
The Gamma variant (P.1) was one of the variants of SARS-CoV-2, the virus that causes COVID-19. This variant of SARS-CoV-2 has been named lineage P.1 and has 17 amino acid substitutions, ten of which in its spike protein, including these three designated to be of particular concern: N501Y, E484K and K417T. This variant of SARS-CoV-2 was first detected by the National Institute of Infectious Diseases (NIID) of Japan, on 6 January 2021 in four people who had arrived in Tokyo having visited Amazonas, Brazil, four days earlier. It was subsequently declared to be in circulation in Brazil. Under the simplified naming scheme proposed by the World Health Organization, P.1 has been labeled Gamma variant, and is currently considered a variant of concern. Gamma caused widespread infection in early 2021 in the city of Manaus, the capital of Amazonas, although the city had already experienced widespread infection in May 2020, with a study indicating high seroprevalence of antibodies for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SARS-CoV-2 Beta Variant
The Beta variant, (B.1.351), was a variant of SARS-CoV-2, the virus that causes COVID-19. One of several SARS-CoV-2 variants initially believed to be of particular importance, it was first detected in the Nelson Mandela Bay metropolitan area of the Eastern Cape province of South Africa in October 2020, which was reported by the country's health department on 18 December 2020. Phylogeographic analysis suggests this variant emerged in the Nelson Mandela Bay area in July or August 2020. The World Health Organization labelled the variant as Beta variant, not to replace the scientific name but as a name for the public to commonly refer to. The WHO considers it to be a variant of concern no longer in circulation. Names The variant is also known as the ''South African variant''. Mutations File:SARS-CoV-2 Beta variant.svg, Amino acid mutations of SARS-CoV-2 Beta variant plotted on a genome map of SARS-CoV-2 with a focus on Spike. There are three mutations of particular interes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SARS-CoV-2 Alpha Variant
The Alpha variant (B.1.1.7) was a Variants of SARS-CoV-2, SARS-CoV-2 variant of concern. It was estimated to be 40–80% more Transmission (medicine), transmissible than the Wild type, wild-type SARS-CoV-2 (with most estimates occupying the middle to higher end of this range). It was first detected in November 2020 from a sample taken in September in the United Kingdom, and began to spread quickly by mid-December, around the same time as infections surged. This increase is thought to be at least partly because of one or more mutations in the virus' coronavirus spike protein, spike protein. The variant was also notable for having more mutations than normally seen. By January 2021, more than half of all genomic sequencing of SARS-CoV-2 was carried out in the UK. This had given rise to questions as to how many other important variants were circulating around the world undetected. On 2 February 2021, Public Health England reported that they had detected "[a] limited number of B ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SARS-CoV-2 Kappa Variant
Kappa variant is a variant of SARS-CoV-2, the virus that causes COVID-19. It is one of the three sublineages of Pango lineage B.1.617. The SARS-CoV-2 Kappa variant is also known as lineage B.1.617.1 and was first detected in India in December 2020. By the end of March 2021, the Kappa sub-variant accounted for more than half of the sequences being submitted from India. On 1 April 2021, it was designated a Variant Under Investigation (VUI-21APR-01) by Public Health England. Mutations File:SARS-CoV-2_Kappa_variant.svg, Amino acid mutations of SARS-CoV-2 Kappa variant plotted on a genome map of SARS-CoV-2 with a focus on the spike. The Kappa variant has three notable alterations in the amino-acid sequences, all of which are in the virus's spike protein code. The three notable substitutions are: L452R, E484Q, P681R * L452R. The substitution at position 452, a leucine-to-arginine substitution. This exchange confers stronger affinity of the spike protein for the ACE2 receptor a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SARS-CoV-2 Iota Variant
Iota variant, also known as lineage B.1.526, is one of the variants of SARS-CoV-2, the virus that causes COVID-19. It was first detected in New York City in November 2020. The variant has appeared with two notable mutations: the E484K spike mutation, which may help the virus evade antibodies, and the S477N mutation, which may help the virus bind more tightly to human cells. By February 2021, it had spread rapidly in the New York region and accounted for about one in four viral sequences. By 11 April 2021, the variant had been detected in at least 48 U.S. states and 18 countries. Under the simplified naming scheme proposed by the World Health Organization, B.1.526 has been labeled Iota variant, and is considered a ''variant of interest'' (VOI), but not yet a variant of concern. Mutations The Iota (B.1.526) genome contains the following amino-acid mutations, all of which are in the virus's spike protein code: L5F, T95I, D253G, E484K, D614G and A701V. File:SARS-CoV-2_Iota_vari ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |