|
RNASEL
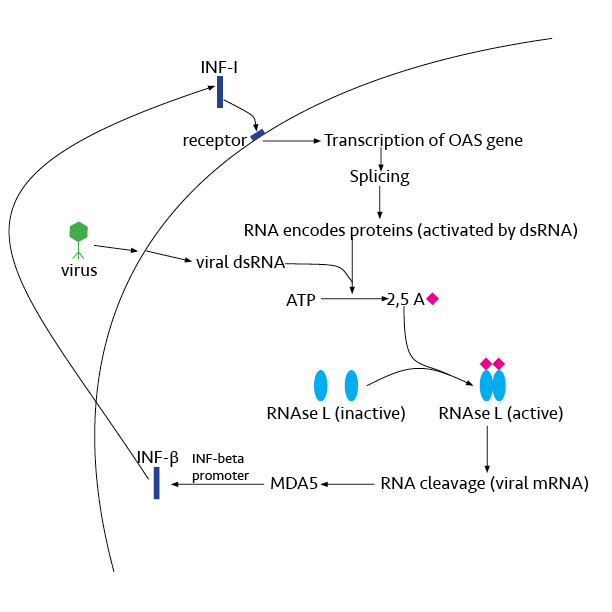
Ribonuclease L or RNase L (for ''latent''), known sometimes as ribonuclease 4 or 2'-5' oligoadenylate synthetase-dependent ribonuclease — is an interferon (IFN)-induced ribonuclease which, upon activation, destroys all RNA within the cell (both cellular and viral). RNase L is an enzyme that in humans is encoded by the ''RNASEL'' gene. This gene encodes a component of the interferon-regulated 2'-5'oligoadenylate (2'-5'A) system that functions in the antiviral and antiproliferative roles of interferons. RNase L is activated by dimerization, which occurs upon 2'-5'A binding, and results in cleavage of all RNA in the cell. This can lead to activation of MDA5, an RNA helicase involved in the production of interferons. Synthesis and activation RNase L is present in very minute quantities during the normal cell cycle. When interferon binds to cell receptors, it activates transcription of around 300 genes to bring about the antiviral state. Among the enzymes produced is RNase L, w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
2'-5'-oligoadenylate Synthase
In molecular biology, 2'-5'-oligoadenylate synthetase (2-5A synthetase) is an enzyme () that reacts to interferon signal. It is an antiviral enzyme that counteracts viral attack by degrading RNAs, both viral and host. The enzyme uses ATP in 2'-specific nucleotidyl transfer reactions to synthesize 2'-5'-oligoadenylates, which activate latent ribonuclease (RNASEL), resulting in degradation of viral RNA and inhibition of virus replication. The C-terminal half of 2'-5'-oligoadenylate synthetase, also referred to as domain 2 of the enzyme, is largely alpha-helical and homologous to a tandem ubiquitin repeat. It carries the region of enzymatic activity between at the extreme C-terminal end. Human proteins * OAS1 * OAS2 * OAS3 2'-5'-oligoadenylate synthetase 3 is an enzyme that in humans is encoded by the ''OAS3'' gene. This gene encodes an enzyme included in the 2', 5' oligoadenylate synthase family. This enzyme is induced by interferons and catalyzes the 2', 5' o ... See al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Interferon Type I
The type-I interferons (IFN) are cytokines which play essential roles in inflammation, immunoregulation, tumor cells recognition, and T-cell responses. In the human genome, a cluster of thirteen functional IFN genes is located at the 9p21.3 cytoband over approximately 400 kb including coding genes for IFNα (''IFNA1, IFNA2, IFNA4, IFNA5, IFNA6, IFNA7, IFNA8, IFNA10, IFNA13, IFNA14, IFNA16, IFNA17'' and ''IFNA21''), IFNω (''IFNW1''), IFNɛ (''IFNE''), IFNк (''IFNK'') and IFNβ (''IFNB1''), plus 11 IFN pseudogenes. Interferons bind to interferon receptors. All type I IFNs bind to a specific cell surface receptor complex known as the IFN-α receptor (IFNAR) that consists of IFNAR1 and IFNAR2 chains. Type I IFNs are found in all mammals, and homologous (similar) molecules have been found in birds, reptiles, amphibians and fish species. Sources and functions IFN-α and IFN-β are secreted by many cell types including lymphocytes (NK cells, B-cells and T-cells), macrophages, fib ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrophosphate
In chemistry, pyrophosphates are phosphorus oxyanions that contain two phosphorus atoms in a P–O–P linkage. A number of pyrophosphate salts exist, such as disodium pyrophosphate (Na2H2P2O7) and tetrasodium pyrophosphate (Na4P2O7), among others. Often pyrophosphates are called diphosphates. The parent pyrophosphates are derived from partial or complete neutralization of pyrophosphoric acid. The pyrophosphate bond is also sometimes referred to as a phosphoanhydride bond, a naming convention which emphasizes the loss of water that occurs when two phosphates form a new P–O–P bond, and which mirrors the nomenclature for anhydrides of carboxylic acids. Pyrophosphates are found in ATP and other nucleotide triphosphates, which are important in biochemistry. The term pyrophosphate is also the name of esters formed by the condensation of a phosphorylated biological compound with inorganic phosphate, as for dimethylallyl pyrophosphate. This bond is also referred to as a high-energy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chronic Fatigue Syndrome
Chronic fatigue syndrome (CFS), also called myalgic encephalomyelitis (ME) or ME/CFS, is a complex, debilitating, long-term medical condition. The causes and mechanisms of the disease are not fully understood. Distinguishing core symptoms are lengthy exacerbations or flare-ups of the illness following ordinary minor physical or mental activity, known as post-exertional malaise (PEM); greatly diminished capacity to accomplish tasks that were routine before the illness; and sleep disturbances. Orthostatic intolerance (difficulty sitting and standing upright) and cognitive dysfunction are also diagnostic. Frequently and variably, other common symptoms occur involving numerous body systems, and chronic pain is common. The unexplained and often incapacitating fatigue in CFS is different from that caused by normal strenuous ongoing exertion, is not significantly relieved by rest, and is not due to a previous medical condition. Diagnosis is based on the person's symptoms because no c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helicase
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands (hence '' helic- + -ase''), using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases. The human genome codes for 95 non-redundant helicases: 64 RNA helicases and 31 DNA helicases. Many cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair, and ribosome biogenesis involve the separation of nucleic acid strands that necessitates the use of helicases. Some specialized helicases are also involved in sensing of viral nucleic acids during infection and fulfill a immunological function. Function Helicases are o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Small Interfering RNA
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA at first non-coding RNA molecules, typically 20-24 (normally 21) base pairs in length, similar to miRNA, and operating within the RNA interference (RNAi) pathway. It interferes with the expression of specific genes with complementary nucleotide sequences by degrading mRNA after transcription, preventing translation. Text was copied from this source, which is available under Creative Commons Attribution 4.0 International License Structure Naturally occurring siRNAs have a well-defined structure that is a short (usually 20 to 24- bp) double-stranded RNA (dsRNA) with phosphorylated 5' ends and hydroxylated 3' ends with two overhanging nucleotides. The Dicer enzyme catalyzes production of siRNAs from long dsRNAs and small hairpin RNAs. siRNAs can also be introduced into cells by transfection. Since in principle any gene can be knocked down by a syntheti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Kinase R
Protein kinase RNA-activated also known as protein kinase R (PKR), interferon-induced, double-stranded RNA-activated protein kinase, or eukaryotic translation initiation factor 2-alpha kinase 2 (EIF2AK2) is an enzyme that in humans is encoded by the ''EIF2AK2'' gene. PKR protects against viral infections. Mechanism of action Protein kinase-R is activated by double-stranded RNA (dsRNA), introduced to the cells by a viral infection. PKR can also be activated by the protein PACT or by heparin. PKR contains an N-terminal dsRNA binding domain (dsRBD) and a C-terminal kinase domain, that gives it pro-apoptotic (cell-killing) functions. The dsRBD consists of two tandem copies of a conserved double stranded RNA binding motif, dsRBM1 and dsRBM2. PKR is induced by interferon in a latent state. Binding to dsRNA is believed to activate PKR by inducing dimerization and subsequent auto-phosphorylation reactions. In situations of viral infection, the dsRNA created by viral replication and g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Förster Resonance Energy Transfer
Förster resonance energy transfer (FRET), fluorescence resonance energy transfer, resonance energy transfer (RET) or electronic energy transfer (EET) is a mechanism describing energy transfer between two light-sensitive molecules ( chromophores). A donor chromophore, initially in its electronic excited state, may transfer energy to an acceptor chromophore through nonradiative dipole–dipole coupling. The efficiency of this energy transfer is inversely proportional to the sixth power of the distance between donor and acceptor, making FRET extremely sensitive to small changes in distance. Measurements of FRET efficiency can be used to determine if two fluorophores are within a certain distance of each other. Such measurements are used as a research tool in fields including biology and chemistry. FRET is analogous to near-field communication, in that the radius of interaction is much smaller than the wavelength of light emitted. In the near-field region, the excited chromophore e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dimer (chemistry)
A dimer () ('' di-'', "two" + ''-mer'', "parts") is an oligomer consisting of two monomers joined by bonds that can be either strong or weak, covalent or intermolecular. Dimers also have significant implications in polymer chemistry, inorganic chemistry, and biochemistry. The term ''homodimer'' is used when the two molecules are identical (e.g. A–A) and ''heterodimer'' when they are not (e.g. A–B). The reverse of dimerization is often called dissociation. When two oppositely charged ions associate into dimers, they are referred to as ''Bjerrum pairs'', after Niels Bjerrum. Noncovalent dimers Anhydrous carboxylic acids form dimers by hydrogen bonding of the acidic hydrogen and the carbonyl oxygen. For example, acetic acid forms a dimer in the gas phase, where the monomer units are held together by hydrogen bonds. Under special conditions, most OH-containing molecules form dimers, e.g. the water dimer. Excimers and exciplexes are excited structures with a short lifetime. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apoptosis
Apoptosis (from grc, ἀπόπτωσις, apóptōsis, 'falling off') is a form of programmed cell death that occurs in multicellular organisms. Biochemical events lead to characteristic cell changes (morphology) and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, DNA fragmentation, and mRNA decay. The average adult human loses between 50 and 70 billion cells each day due to apoptosis. For an average human child between eight and fourteen years old, approximately twenty to thirty billion cells die per day. In contrast to necrosis, which is a form of traumatic cell death that results from acute cellular injury, apoptosis is a highly regulated and controlled process that confers advantages during an organism's life cycle. For example, the separation of fingers and toes in a developing human embryo occurs because cells between the digits undergo apoptosis. Unlike necrosis, apoptosis produces cell fragments called apoptotic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autophagy
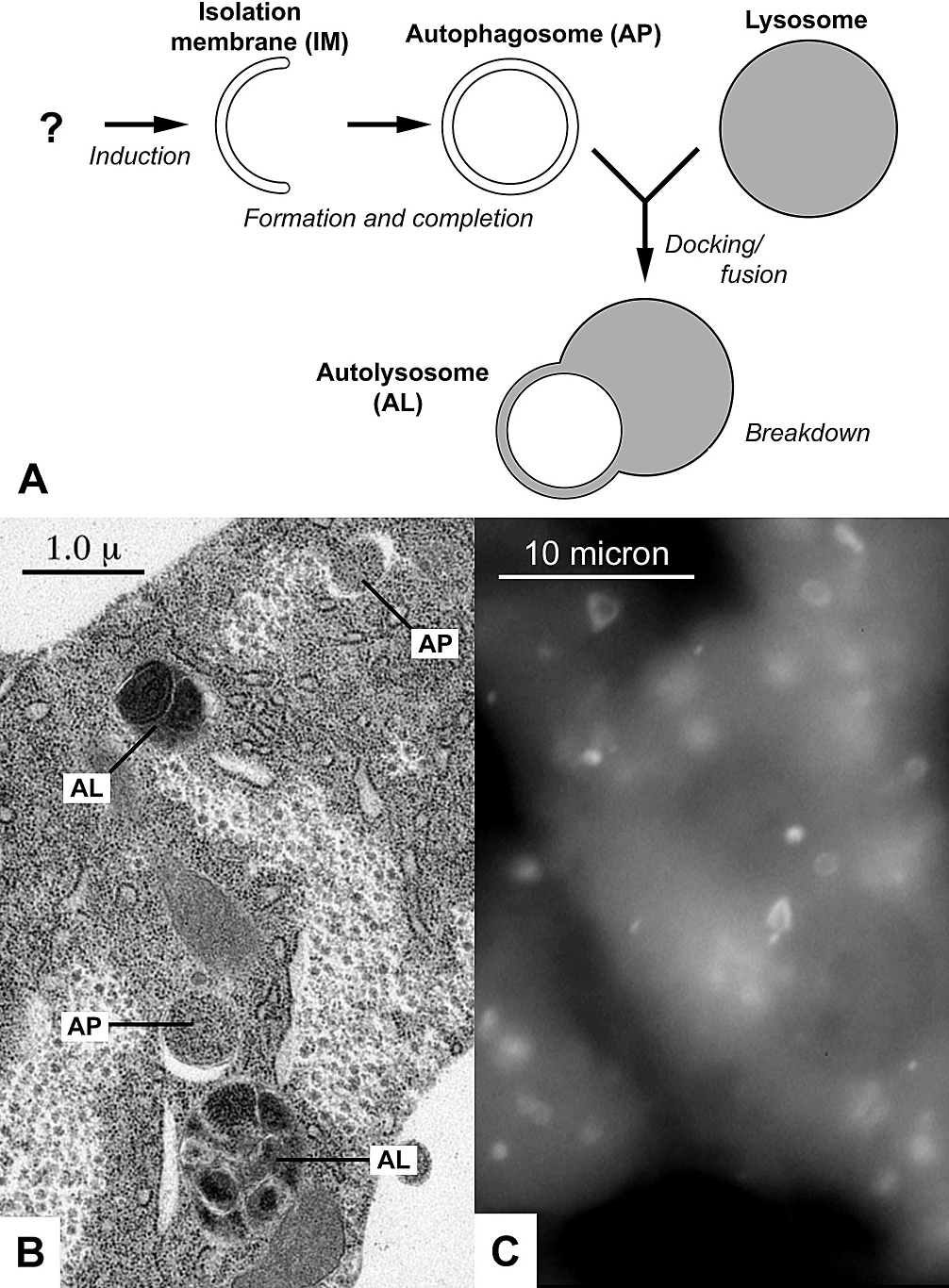
Autophagy (or autophagocytosis; from the Ancient Greek , , meaning "self-devouring" and , , meaning "hollow") is the natural, conserved degradation of the cell that removes unnecessary or dysfunctional components through a lysosome-dependent regulated mechanism. It allows the orderly degradation and recycling of cellular components. Although initially characterized as a primordial degradation pathway induced to protect against starvation, it has become increasingly clear that autophagy also plays a major role in the homeostasis of non-starved cells. Defects in autophagy have been linked to various human diseases, including neurodegeneration and cancer, and interest in modulating autophagy as a potential treatment for these diseases has grown rapidly. Four forms of autophagy have been identified: macroautophagy, microautophagy, chaperone-mediated autophagy (CMA), and crinophagy. In macroautophagy (the most thoroughly researched form of autophagy), cytoplasmic components (like mit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complementary DNA
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), or to sequence or quantify mRNA molecules using DNA based methods (qPCR, RNA-seq). cDNA that codes for a specific protein can be transferred to a recipient cell for expression, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays, qPCR, and RNA-seq. cDNA is also produced naturally by retroviruses (such as HIV-1, HIV-2, simian immunodeficiency virus, etc.) and then integrated into the host's genome, where it creates a provirus. The term ''cDNA'' is also used, typically in a bioinformatics context, to refer to a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |