|
Pseudomonas SRNA P26
''Pseudomonas'' sRNA P26 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen '' Pseudomonas aeruginosa'' and its expression verified by northern blot analysis. P26 is conserved across many Gammaproteobacteria species and appears to be consistently located between the DNA directed RNA polymerase (beta subunit) and 50S ribosomal protein L7/L12 genes. See also *Pseudomonas sRNA P9 * Pseudomonas sRNA P11 *Pseudomonas sRNA P15 *Pseudomonas sRNA P16 *Pseudomonas sRNA P24 *Pseudomonas sRNA P1 ''Pseudomonas'' sRNA P1 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen ''Pseudomonas aeruginosa'' and its expression verified by northern blot analysis. There appears to be two related copies ... References External links * Non-coding RNA {{molecular-cell-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protein coding, i.e. messenger RNA (mRNA); or n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudomonas SRNA P24
''Pseudomonas'' sRNA P24 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen ''Pseudomonas aeruginosa'' and its expression verified by northern blot analysis. P24 is conserved across several ''Pseudomonas'' species and is consistently located between a hypothetical protein gene and a transcriptional regulator gene (AsnC family) in the genomes of these ''Pseudomonas'' species. P24 has a predicted Rho independent terminatorat the 3′ end but the function of P24 is unknown. See also * Pseudomonas sRNA P9 *Pseudomonas sRNA P11 *Pseudomonas sRNA P15 Pseudomonas sRNA P15 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen ''Pseudomonas aeruginosa'' and its expression verified by northern blot analysis. P15 is conserved across several ''Pseudomo ... * Pseudomonas sRNA P16 * Pseudomonas sRNA P26 References External links * Non-coding RNA {{molecular-cell-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudomonas SRNA P16
''Pseudomonas'' sRNA P16 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen ''Pseudomonas aeruginosa'' and its expression verified by northern blot analysis. P16 sRNA appears to be conserved across several ''Pseudomonas'' species and is consistently located downstream of a predicted TatD deoxyribonuclease gene. P16 has a predicted Rho independent terminator at the 3′ end but the function of P16 is unknown. It has been shown that this sRNA is transcribed from an RpoS-dependent promoter under positive, probably indirect GacA control in two ''Pseudomonas'' species. It was renamed RgsA (for regulation by GacA and stress). RpoS mRNA expression is repressed by RgsA during the exponential phase. The Hfq RNA chaperone is required for the repression. See also * Pseudomonas sRNA P1 * Pseudomonas sRNA P9 *Pseudomonas sRNA P11 *Pseudomonas sRNA P15 *Pseudomonas sRNA P24 ''Pseudomonas'' sRNA P24 is a ncRNA that was predicted using bioinf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudomonas SRNA P15
Pseudomonas sRNA P15 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen ''Pseudomonas aeruginosa'' and its expression verified by northern blot analysis. P15 is conserved across several ''Pseudomonas'' species and is consistently located upstream of a 3-deoxy-7-phosphoheptulonate synthase gene. P15 has a predicted Rho independent terminator at the 3′ end but the function of P15 is unknown. See also * Pseudomonas sRNA P1 * Pseudomonas sRNA P9 *Pseudomonas sRNA P11 ''Pseudomonas'' sRNA P11 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen ''Pseudomonas aeruginosa'' and its expression verified by northern blot analysis. P11 is located between a putative thre ... * Pseudomonas sRNA P16 * Pseudomonas sRNA P24 * Pseudomonas sRNA P26 References External links * Non-coding RNA {{molecular-cell-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudomonas SRNA P11
''Pseudomonas'' sRNA P11 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen '' Pseudomonas aeruginosa'' and its expression verified by northern blot analysis. P11 is located between a putative threonine protein kinase and putative nitrate reductase and is conserved in several '' Pseudomonas'' species. P11 has a predicted Rho independent terminator at the 3′ end but the function of P11 is unknown. See also *Pseudomonas sRNA P1 *Pseudomonas sRNA P9 *Pseudomonas sRNA P15 *Pseudomonas sRNA P16 *Pseudomonas sRNA P24 ''Pseudomonas'' sRNA P24 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen ''Pseudomonas aeruginosa'' and its expression verified by northern blot analysis. P24 is conserved across several ''Pseu ... * Pseudomonas sRNA P26 References External links * Non-coding RNA {{molecular-cell-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudomonas SRNA P9
''Pseudomonas'' sRNA P9 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen ''Pseudomonas aeruginosa'' and its expression verified by northern blot analysis. P9 appears to be conserved in several ''Pseudomonas'' species in addition to ''Bordetella'' species. In both ''Pseudomonas'' and ''Bordetella'' species P9 appears to be located upstream of a predicted threonine dehydratase gene. P9 has a predicted Rho independent terminator at the 3′ Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ri ... end but the function of P9 is unknown. See also * Pseudomonas sRNA P1 *Pseudomonas sRNA P11 *Pseudomonas sRNA P15 *Pseudomonas sRNA P16 *Pseudomonas sRNA P24 *Pseudomonas sRNA P26 References External links * Non-coding RNA {{molecular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genes
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins (RPs or r-proteins). The ribosomes and associated molecules are also known as the ''translational apparatus''. Overview The sequence of DNA that encodes the sequence of the amino acids in a protein is transcribed into a messenger RNA chain. Ribosomes bind to messenger RNAs and use their sequences for determining the correct sequence of amino acids to generate a given protein. Amino acids are selected and carried to the ribosome by transfer RNA (tRNA) molecules, which enter the ribosome and bind to the messenger RNA chain via an anti-codon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gammaproteobacteria
Gammaproteobacteria is a class of bacteria in the phylum Pseudomonadota (synonym Proteobacteria). It contains about 250 genera, which makes it the most genera-rich taxon of the Prokaryotes. Several medically, ecologically, and scientifically important groups of bacteria belong to this class. It is composed by all Gram-negative microbes and is the most phylogenetically and physiologically diverse class of Proteobacteria. These microorganisms can live in several terrestrial and marine environments, in which they play various important roles, including ''extreme environments'' such as hydrothermal vents. They generally have different shapes - rods, curved rods, cocci, spirilla, and filaments and include free living bacteria, biofilm formers, commensals and symbionts, some also have the distinctive trait of being bioluminescent. Metabolisms found in the different genera are very different; there are both aerobic and anaerobic (obligate or facultative) species, chemolithoautotrophic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Conservation
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, promp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
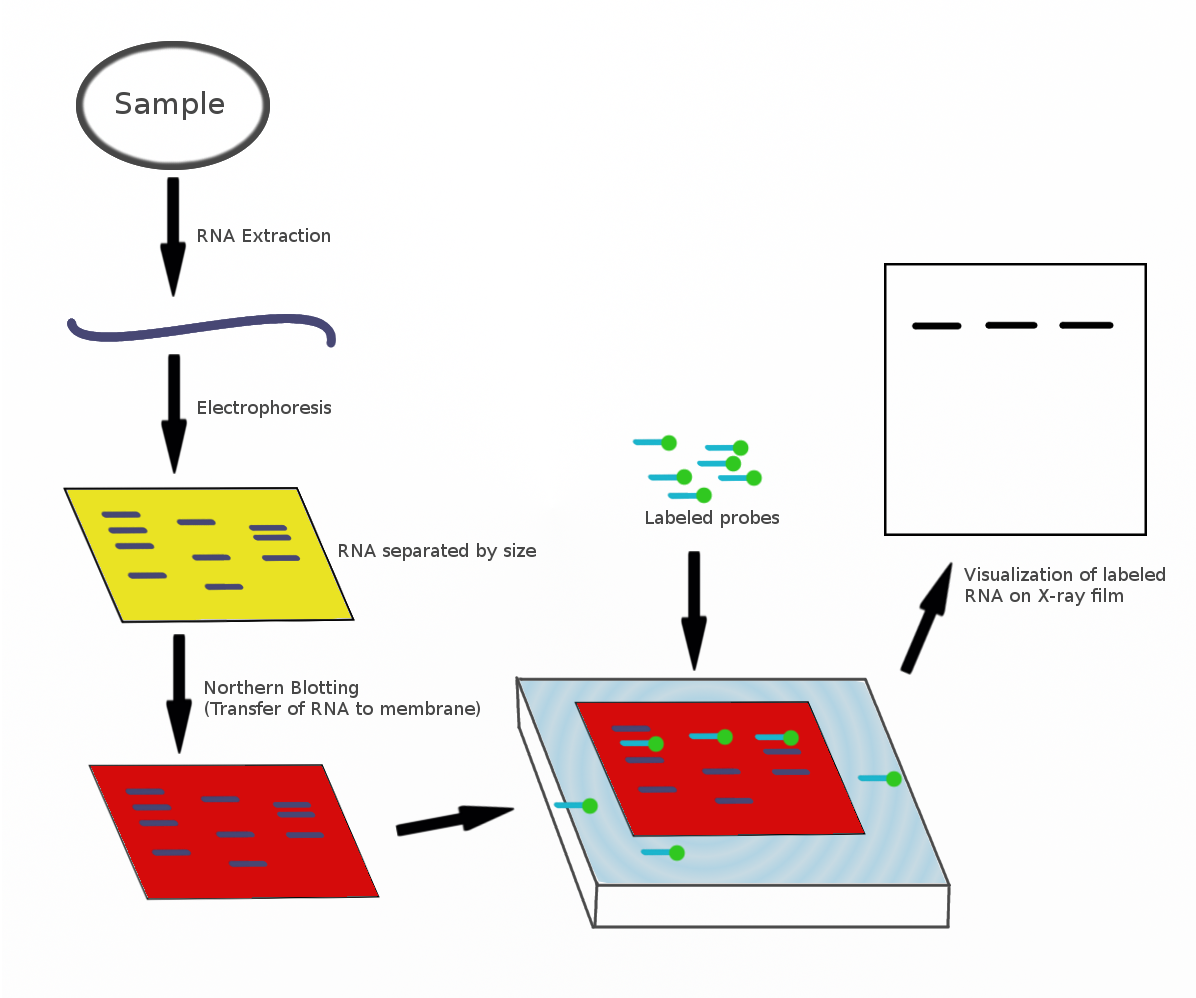
Northern Blot
The northern blot, or RNA blot,Gilbert, S. F. (2000) Developmental Biology, 6th Ed. Sunderland MA, Sinauer Associates. is a technique used in molecular biology research to study gene expression by detection of RNA (or isolated mRNA) in a sample.Kevil, C. G., Walsh, L., Laroux, F. S., Kalogeris, T., Grisham, M. B., Alexander, J. S. (1997) An Improved, Rapid Northern Protocol. Biochem. and Biophys. Research Comm. 238:277–279. With northern blotting it is possible to observe cellular control over structure and function by determining the particular gene expression rates during differentiation and morphogenesis, as well as in abnormal or diseased conditions. Northern blotting involves the use of electrophoresis to separate RNA samples by size, and detection with a hybridization probe complementary to part of or the entire target sequence. Strictly speaking, the term 'northern blot' refers specifically to the capillary transfer of RNA from the electrophoresis gel to the blotting m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |