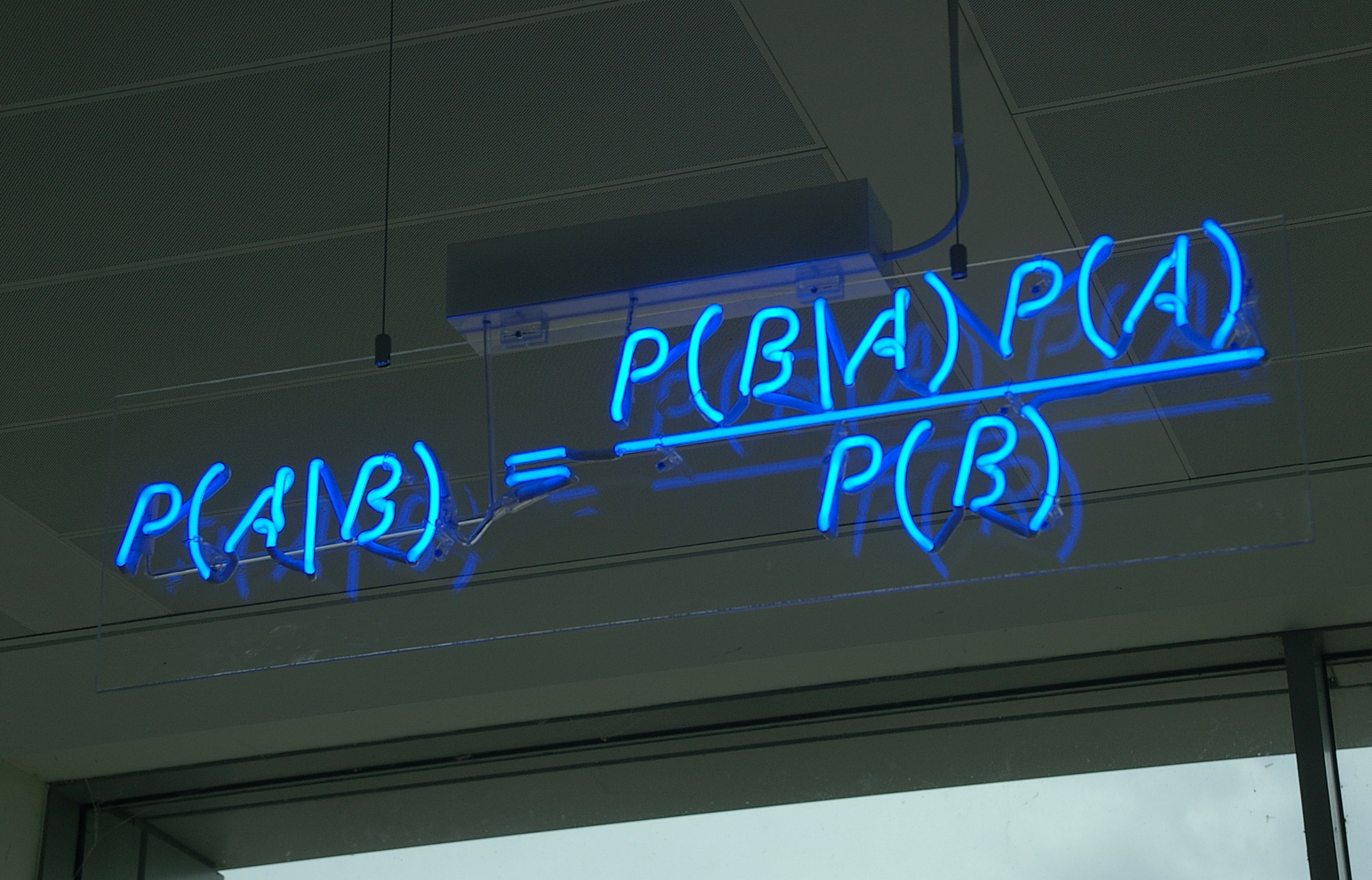|
Phylogenetics Software
This list of phylogenetics software is a compilation of computational phylogenetics software used to produce phylogenetic trees. Such tools are commonly used in comparative genomics, cladistics, and bioinformatics. Methods for estimating phylogenies include neighbor-joining, maximum parsimony (also simply referred to as parsimony), UPGMA, Bayesian phylogenetic inference, maximum likelihood and distance matrix methods. List See also *List of phylogenetic tree visualization software References External links *Complete list oInstitut Pasteurphylogeny webserversExPASyList of phylogenetics programs *A verof phylogenetic tools (reconstruction, visualization, ''etc.'') *Anothelist of evolutionary genetics software*A list ophylogenetic softwareprovided by the Zoological Research Museum A. Koenig *MicrobeTrace available at https://github.com/CDCgov/MicrobeTrace/wiki {{DEFAULTSORT:List Of Phylogenetics Software Genetics databases Phylo Phylogenetics In biology, phylo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Computational Phylogenetics
Computational phylogenetics is the application of computational algorithms, methods, and programs to phylogenetic"origin,_source,_birth")_is_the_study_of_the_evolutionary_his_... "origin, source, birth") is the study of the evolutionary his ... "origin, source, birth") is the study of the evolutionary his ... "origin, source, birth") is the study of the evolutionary his ... "origin, source, birth") is the study of the evolutionary his ... "tribe, clan, race", and wikt:γενετικός, γενετικός [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Markov Chain Monte Carlo
In statistics, Markov chain Monte Carlo (MCMC) methods comprise a class of algorithms for sampling from a probability distribution. By constructing a Markov chain that has the desired distribution as its equilibrium distribution, one can obtain a sample of the desired distribution by recording states from the chain. The more steps that are included, the more closely the distribution of the sample matches the actual desired distribution. Various algorithms exist for constructing chains, including the Metropolis–Hastings algorithm. Application domains MCMC methods are primarily used for calculating numerical approximations of multi-dimensional integrals, for example in Bayesian statistics, computational physics, computational biology and computational linguistics. In Bayesian statistics, the recent development of MCMC methods has made it possible to compute large hierarchical models that require integrations over hundreds to thousands of unknown parameters. In rare even ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PAUP*
PAUP* (Phylogenetic Analysis Using Parsimony *and other methods) is a computational phylogenetics program for inferring evolutionary trees (phylogenies), written by David L. Swofford. Originally, as the name implies, PAUP only implemented parsimony, but from version 4.0 (when the program became known as PAUP*) it also supports distance matrix and likelihood methods. Version 3.0 ran on Macintosh computers and supported a rich, user-friendly graphical interface. Together with the program MacClade, with which it shares the NEXUS data format, PAUP* was the phylogenetic software of choice for many phylogenetists. Version 4.0 added support for Windows (graphical shell and command line) and Unix (command line only) platforms. However, the graphical user interface for the Macintosh does not support versions of Mac OS X higher than 10.14 (although a GUI for later versions of Mac OS is planned). PAUP* is also available as a plugin for Geneious. PAUP*, which now sports the self-referent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ziheng Yang
Ziheng Yang FRS (; born 1 November 1964) is a Chinese biologist. He holds the R.A. Fisher Chair of Statistical Genetics at University College London,‘YANG, Prof. Ziheng’, Who's Who 2011, A & C Black, 2011; online edn, Oxford University Press, Dec 2010 ; online edn, Oct 201accessed 11 May 2011/ref> and is the Director of R.A. Fisher Centre for Computational Biology at UCL. He was elected a Fellow of the Royal Society in 2006. Academic career Yang graduated from Gansu Agricultural University with a BSc in 1984, and from Beijing Agricultural University with a MSc in 1987, and PhD in 1992. After the PhD, he worked as a postdoctoral researcher in Department of Zoology, University of Cambridge (1992-3), The Natural History Museum (London) (1993-4), Pennsylvania State University (1994-5), and University of California at Berkeley (1995-7), before taking up a faculty position in Department of Biology, University College London. He was a Lecturer (1997), Reader (2000), and then P ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bayesian Inference
Bayesian inference is a method of statistical inference in which Bayes' theorem is used to update the probability for a hypothesis as more evidence or information becomes available. Bayesian inference is an important technique in statistics, and especially in mathematical statistics. Bayesian updating is particularly important in the dynamic analysis of a sequence of data. Bayesian inference has found application in a wide range of activities, including science, engineering, philosophy, medicine, sport, and law. In the philosophy of decision theory, Bayesian inference is closely related to subjective probability, often called "Bayesian probability". Introduction to Bayes' rule Formal explanation Bayesian inference derives the posterior probability as a consequence of two antecedents: a prior probability and a "likelihood function" derived from a statistical model for the observed data. Bayesian inference computes the posterior probability according to Bayes' theorem: ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MrBayes
Bayesian inference of phylogeny combines the information in the prior and in the data likelihood to create the so-called posterior probability of trees, which is the probability that the tree is correct given the data, the prior and the likelihood model. Bayesian inference was introduced into molecular phylogenetics in the 1990s by three independent groups: Bruce Rannala and Ziheng Yang in Berkeley, Bob Mau in Madison, and Shuying Li in University of Iowa, the last two being PhD students at the time. The approach has become very popular since the release of the MrBayes software in 2001, and is now one of the most popular methods in molecular phylogenetics. Bayesian inference of phylogeny background and bases Bayesian inference refers to a probabilistic method developed by Reverend Thomas Bayes based on Bayes' theorem. Published posthumously in 1763 it was the first expression of inverse probability and the basis of Bayesian inference. Independently, unaware of Bayes' work, Pierre- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Algorithm
In computer science and operations research, a genetic algorithm (GA) is a metaheuristic inspired by the process of natural selection that belongs to the larger class of evolutionary algorithms (EA). Genetic algorithms are commonly used to generate high-quality solutions to optimization and search problems by relying on biologically inspired operators such as mutation, crossover and selection. Some examples of GA applications include optimizing decision trees for better performance, solving sudoku puzzles, hyperparameter optimization, etc. Methodology Optimization problems In a genetic algorithm, a population of candidate solutions (called individuals, creatures, organisms, or phenotypes) to an optimization problem is evolved toward better solutions. Each candidate solution has a set of properties (its chromosomes or genotype) which can be mutated and altered; traditionally, solutions are represented in binary as strings of 0s and 1s, but other encodings are also possible. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Wayne Maddison
Wayne Paul Maddison , is a professor and Canada Research Chair at the departments of zoology and botany at the University of British Columbia, and the Director of the Spencer Entomological Collection at the Beaty Biodiversity Museum. His research concerns the phylogeny, biodiversity, and evolution of jumping spiders (Salticidae), of which he has discovered new species and genera. He has also done research in phylogenetic theory, developing and perfecting various methods used in comparative biology, such as character state inference in internal nodes through Maximum parsimony (phylogenetics), maximum parsimony, squared-change parsimony, or character correlation through the concentrated changes test or pairwise comparisons. In collaboration with David R. Maddison, he worked on thMesquiteopen-source phylogeny software, thMacCladeprogram, and the Tree of Life Web Project. His research has led him to discover new species of jumping spiders in Sarawak and Papua New Guinea. Selected pu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNASTAR
DNASTAR is a global bioinformatics software company incorporated in 1984 that is headquartered in Madison, Wisconsin. DNASTAR develops and sells software for sequence analysis in the fields of genomics, molecular biology, and structural biology. Software DNASTAR software (Lasergene) first gained popularity in the 1980s and 1990s for its sequence assembly and analysis capabilities of Sanger sequencing data. Lasergene 17.3.3 was released in June 2022. DNASTAR software is available for desktop computers running Mac OS X, Windows, and Linux as well as for use on Amazon Web Services. In 2007, DNASTAR expanded their offerings to include software for next-generation sequencing and structural biology. DNASTAR's next-gen software supports data from Illumina, Ion Torrent, and Pacific Biosciences and allows the user to assemble, align, analyze and visualize genomic data. Lasergene's use in next-generation sequence assembly and analysis was contributed as a chapter, written by company sci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MEGA, Molecular Evolutionary Genetics Analysis
Molecular Evolutionary Genetics Analysis (MEGA) is computer software for conducting statistical analysis of molecular evolution and for constructing phylogenetic trees. It includes many sophisticated methods and tools for phylogenomics and phylomedicine. It is licensed as proprietary freeware. The project for developing this software was initiated by the leadership of Masatoshi Nei in his laboratory at the Pennsylvania State University in collaboration with his graduate student Sudhir Kumar and postdoctoral fellow Koichiro Tamura.Kumar, S., K. Tamura, and M. Nei (1993) MEGA: Molecular Evolutionary Genetics Analysis. Ver. 1.0, The Pennsylvania State University, University Park, PA. Nei wrote a monograph (pp. 130) outlining the scope of the software and presenting new statistical methods that were included in MEGA. The entire set of computer programs was written by Kumar and Tamura. The personal computers then lacked the ability to send the monograph and software electronicall ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MinHash
In computer science and data mining, MinHash (or the min-wise independent permutations locality sensitive hashing scheme) is a technique for quickly estimating how similar two sets are. The scheme was invented by , and initially used in the AltaVista search engine to detect duplicate web pages and eliminate them from search results.. It has also been applied in large-scale clustering problems, such as clustering documents by the similarity of their sets of words.. Jaccard similarity and minimum hash values The Jaccard similarity coefficient is a commonly used indicator of the similarity between two sets. Let be a set and and be subsets of , then the Jaccard index is defined to be the ratio of the number of elements of their intersection and the number of elements of their union: : J(A,B) = . This value is 0 when the two sets are disjoint, 1 when they are equal, and strictly between 0 and 1 otherwise. Two sets are more similar (i.e. have relatively more members in common) wh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HYPHY (software)
HYPHY ( ) is a free multiplatform (Mac, Windows and UNIX) computational phylogenetics software package intended to perform maximum likelihood analyses of genetic sequence data and equipped with tools to test various statistical hypotheses. The HYPHY name is an abbreviation for "HYpothesis testing using PHYlogenies". As of March 2018, about 2,000 peer-reviewed scientific journal articles cite HYPHY. Major features HYPHY supports analysis of nucleotide, protein and codon sequences, using predefined standard models or user-defined models of evolution. The package supports interaction through a graphical user interface as well as a batch language to set up large and complicated analyses and process the results. HyPhy includes a versatile suite of methods to detect adaptive evolution at individual amino-acid sites and/or lineages, including generalizations of Nielsen-Yang PAML and Suzuki-Gojobori approaches and many others. History The development of HyPhy started in 1997, wit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |