|
Phagemid
A phagemid or phasmid is a DNA-based cloning vector, which has both bacteriophage and plasmid properties. These vectors carry, in addition to the origin of plasmid replication, an origin of replication derived from bacteriophage. Unlike commonly used plasmids, phagemid vectors differ by having the ability to be packaged into the capsid of a bacteriophage, due to their having a genetic sequence that signals for packaging. Phagemids are used in a variety of biotechnology applications; for example, they can be used in a molecular biology technique called "Phage Display". Properties of the cloning vector A phagemid (plasmid + phage) is a plasmid that contains an f1 origin of replication from an f1 phage.Analysis of Genes and Genomes, John Wiley & Sons, 2004, S. 140Google Books/ref> It can be used as a type of cloning vector in combination with filamentous phage M13. A phagemid can be replicated as a plasmid, and also be packaged as single stranded DNA in viral particles. Phagemids ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phage Display
Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein– DNA interactions that uses bacteriophages (viruses that infect bacteria) to connect proteins with the genetic information that encodes them. In this technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to "display" the protein on its outside while containing the gene for the protein on its inside, resulting in a connection between genotype and phenotype. These displaying phages can then be screened against other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. In this way, large libraries of proteins can be screened and amplified in a process called ''in vitro'' selection, which is analogous to natural selection. The most common bacteriophages used in phage display are M13 and fd filamentous phage, though T4, T7, and λ phage have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phage Display
Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein– DNA interactions that uses bacteriophages (viruses that infect bacteria) to connect proteins with the genetic information that encodes them. In this technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to "display" the protein on its outside while containing the gene for the protein on its inside, resulting in a connection between genotype and phenotype. These displaying phages can then be screened against other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. In this way, large libraries of proteins can be screened and amplified in a process called ''in vitro'' selection, which is analogous to natural selection. The most common bacteriophages used in phage display are M13 and fd filamentous phage, though T4, T7, and λ phage have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cloning Vector
A cloning vector is a small piece of DNA that can be stably maintained in an organism, and into which a foreign DNA fragment can be inserted for cloning purposes. The cloning vector may be DNA taken from a virus, the cell of a higher organism, or it may be the plasmid of a bacterium. The vector contains features that allow for the convenient insertion of a DNA fragment into the vector or its removal from the vector, for example through the presence of restriction sites. The vector and the foreign DNA may be treated with a restriction enzyme that cuts the DNA, and DNA fragments thus generated contain either blunt ends or overhangs known as sticky ends, and vector DNA and foreign DNA with compatible ends can then be joined together by molecular ligation. After a DNA fragment has been cloned into a cloning vector, it may be further subcloned into another vector designed for more specific use. There are many types of cloning vectors, but the most commonly used ones are genetically ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Site-directed Mutagenesis
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional mutating changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenesis, it is used for investigating the structure and biological activity of DNA, RNA, and protein molecules, and for protein engineering. Site-directed mutagenesis is one of the most important laboratory techniques for creating DNA libraries by introducing mutations into DNA sequences. There are numerous methods for achieving site-directed mutagenesis, but with decreasing costs of oligonucleotide synthesis, artificial gene synthesis is now occasionally used as an alternative to site-directed mutagenesis. Since 2013, the development of the CRISPR/Cas9 technology, based on a prokaryotic viral defense system, has also allowed for the editing of the genome, and mutagenesis may be performed ''in vivo'' with relative ease. History Early attempt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electroporation
Electroporation, or electropermeabilization, is a microbiology technique in which an electrical field is applied to cells in order to increase the permeability of the cell membrane, allowing chemicals, drugs, electrode arrays or DNA to be introduced into the cell (also called electrotransfer). In microbiology, the process of electroporation is often used to transform bacteria, yeast, or plant protoplasts by introducing new coding DNA. If bacteria and plasmids are mixed together, the plasmids can be transferred into the bacteria after electroporation, though depending on what is being transferred, cell-penetrating peptides or CellSqueeze could also be used. Electroporation works by passing thousands of volts (~8 kV/cm) across suspended cells in an electroporation cuvette. Afterwards, the cells have to be handled carefully until they have had a chance to divide, producing new cells that contain reproduced plasmids. This process is approximately ten times more effective in increasing ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
F-plasmid
The fertility factor (first named F by one of its discoverers Esther Lederberg; also called the sex factor in '' E. coli'' or the F sex factor; also called F-plasmid) allows genes to be transferred from one bacterium carrying the factor to another bacterium lacking the factor by conjugation. The F factor was the first plasmid to be discovered. Unlike other plasmids, F factor is constitutive for transfer proteins due to a mutation in the gene ''finO''. The F plasmid belongs to a class of conjugative plasmids that control sexual functions of bacteria with a fertility inhibition (Fin) system. Discovery Esther M. Lederberg and Luigi L. Cavalli-Sforza discovered "F," subsequently publishing with Joshua Lederberg. Once her results were announced, two other labs joined the studies. "This was not a simultaneous independent discovery of F (I names as Fertility Factor until it was understood.) We wrote to Hayes, Jacob, & Wollman who then proceeded with their studies." The discovery of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pilus
A pilus (Latin for 'hair'; plural: ''pili'') is a hair-like appendage found on the surface of many bacteria and archaea. The terms ''pilus'' and '' fimbria'' (Latin for 'fringe'; plural: ''fimbriae'') can be used interchangeably, although some researchers reserve the term ''pilus'' for the appendage required for bacterial conjugation. All conjugative pili are primarily composed of pilin – fibrous proteins, which are oligomeric. Dozens of these structures can exist on the bacterial and archaeal surface. Some bacteria, viruses or bacteriophages attach to receptors on pili at the start of their reproductive cycle. Pili are antigenic. They are also fragile and constantly replaced, sometimes with pili of different composition, resulting in altered antigenicity. Specific host responses to old pili structures are not effective on the new structure. Recombination genes of pili code for variable (V) and constant (C) regions of the pili (similar to immunoglobulin diversity). As the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysis
Lysis ( ) is the breaking down of the membrane of a cell, often by viral, enzymic, or osmotic (that is, "lytic" ) mechanisms that compromise its integrity. A fluid containing the contents of lysed cells is called a ''lysate''. In molecular biology, biochemistry, and cell biology laboratories, cell cultures may be subjected to lysis in the process of purifying their components, as in protein purification, DNA extraction, RNA extraction, or in purifying organelles. Many species of bacteria are subject to lysis by the enzyme lysozyme, found in animal saliva, egg white, and other secretions. Phage lytic enzymes (lysins) produced during bacteriophage infection are responsible for the ability of these viruses to lyse bacterial cells. Penicillin and related β-lactam antibiotics cause the death of bacteria through enzyme-mediated lysis that occurs after the drug causes the bacterium to form a defective cell wall. If the cell wall is completely lost and the penicillin was used ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
T7 Phage
Bacteriophage T7 (or the T7 phage) is a bacteriophage, a virus that infects bacteria. It infects most strains of ''Escherichia coli'' and relies on these hosts to propagate. Bacteriophage T7 has a lytic life cycle, meaning that it destroys the cell it infects. It also possesses several properties that make it an ideal phage for experimentation: its purification and concentration have produced consistent values in chemical analyses; it can be rendered noninfectious by exposure to UV light; and it can be used in phage display to clone RNA binding proteins. Discovery In a 1945 study by Demerec and Fano, T7 was used to describe one of the seven phage types (T1 to T7) that grow lytically on ''Escherichia coli;'' although all seven phages were numbered arbitrarily, phages with odd numbers, or T-odd phages, were later discovered to share morphological and biochemical features that distinguish them from T-even phages. Before being physically referred to as T7, the phage was used in p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lambda Phage
''Enterobacteria phage λ'' (lambda phage, coliphage λ, officially ''Escherichia virus Lambda'') is a bacterial virus, or bacteriophage, that infects the bacterial species ''Escherichia coli'' (''E. coli''). It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell. The phage particle consists of a head (also known as a capsid), a tail, and tail fibers (see image of virus below). The head contains the phage's double-strand linear DNA genome. During infection, the phage particle recognizes and binds to its host, ''E. coli'', causing DNA in the head of the phage to be ejected through the tail into the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
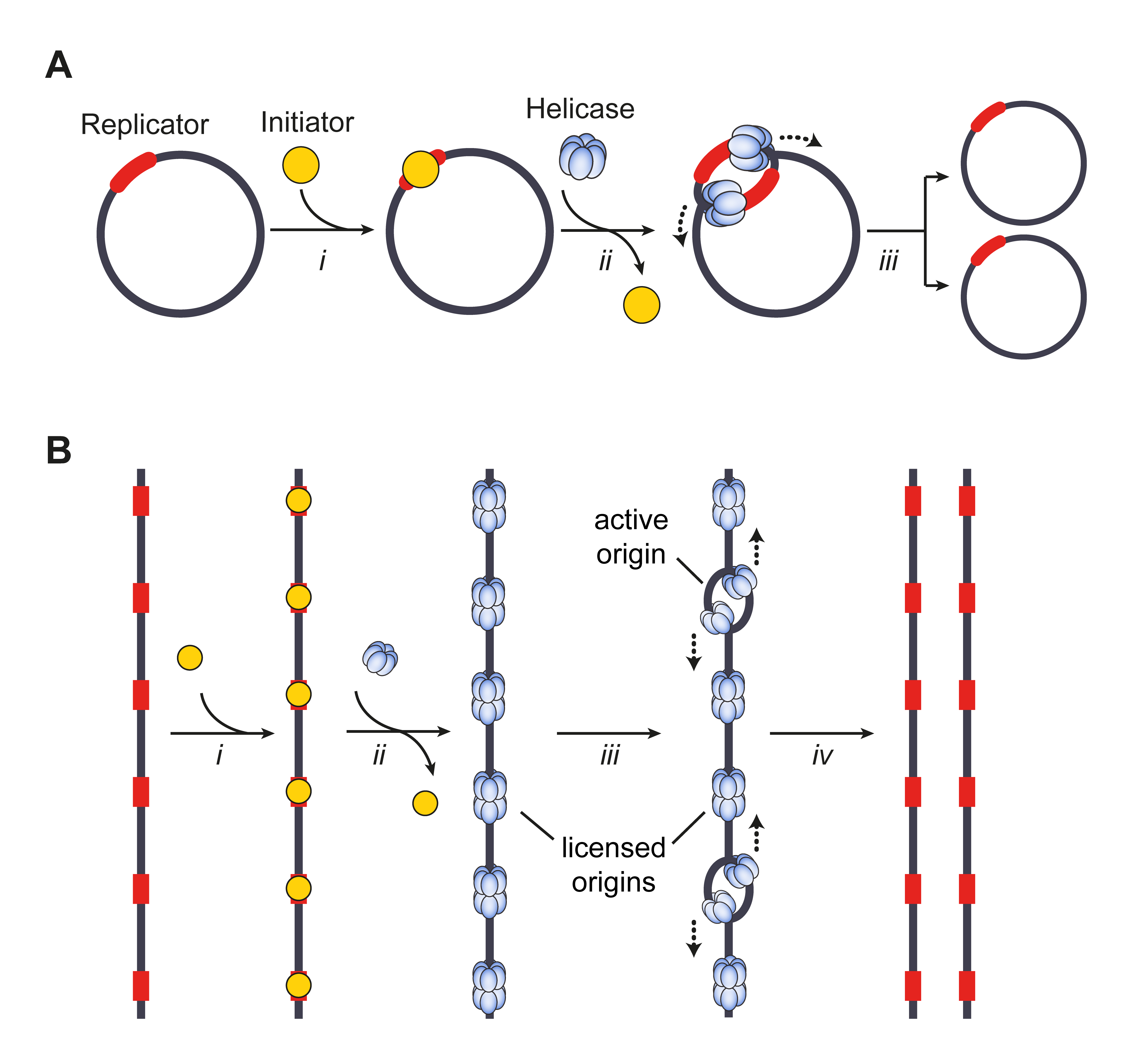
Ori (genetics)
The origin of replication (also called the replication origin) is a particular sequence in a genome at which replication is initiated. Propagation of the genetic material between generations requires timely and accurate duplication of DNA by semiconservative replication prior to cell division to ensure each daughter cell receives the full complement of chromosomes. Material was copied from this source, which is available under Creative Commons Attribution 4.0 International License This can either involve the replication of DNA in living organisms such as prokaryotes and eukaryotes, or that of DNA or RNA in viruses, such as double-stranded RNA viruses. Synthesis of daughter strands starts at discrete sites, termed replication origins, and proceeds in a bidirectional manner until all genomic DNA is replicated. Despite the fundamental nature of these events, organisms have evolved surprisingly divergent strategies that control replication onset. Although the specific replication or ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |