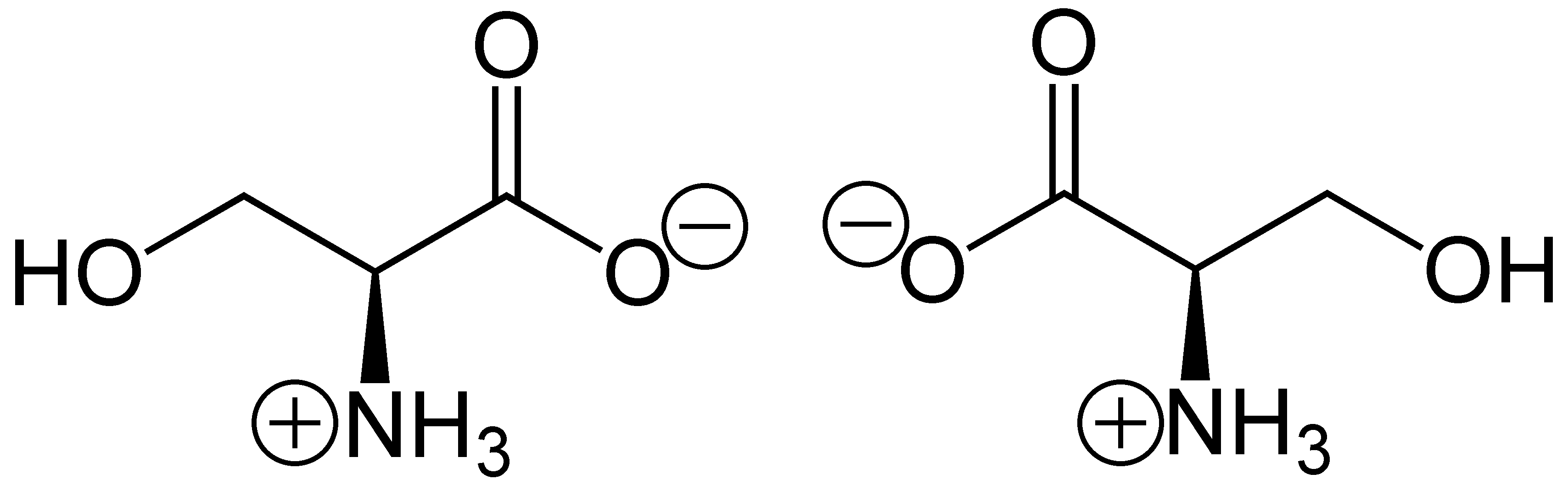|
PETase
PETases are an esterase class of enzymes that catalyze the hydrolysis of polyethylene terephthalate (PET) plastic to monomeric mono-2-hydroxyethyl terephthalate (MHET). The idealized chemical reaction is (where n is the number of monomers in the polymer chain): :(ethylene terephthalate)n + H2O → (ethylene terephthalate)n-1 + MHET Trace amount of the PET breaks down to bis(2-hydroxyethyl) terephthalate (BHET). PETases can also break down PEF-plastic ( polyethylene-2,5-furandicarboxylate), which is a bioderived PET replacement, into the analogous . PETases can't catalyze the hydrolysis of aliphatic polyesters like polybutylene succinate or polylactic acid. Non-enzymatic natural degradation of PET will take hundreds of years, but PETases can degrade PET in matter of days. History The first PETase was discovered in 2016 from ''Ideonella sakaiensis'' strain 201-F6 bacteria found from sludge samples collected close to a Japanese PET bottle recycling site. Other types of PET degradi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ideonella Sakaiensis
''Ideonella sakaiensis'' is a bacterium from the genus'' Ideonella'' and family Comamonadaceae capable of breaking down and consuming the plastic polyethylene terephthalate (PET) using it as both a carbon and energy source. The bacterium was originally isolated from a sediment sample taken outside of a plastic bottle recycling facility in Sakai City, Japan. * Discovery ''Ideonella sakaiensis'' was first identified in 2016 by a team of researchers led by Kohei Oda of Kyoto Institute of Technology and Kenji Miyamoto of Keio University after collecting a sample of PET-contaminated sediment at a plastic bottle recycling facility in Sakai, Japan. The bacteria was first isolated from a consortium of microorganisms in the sediment sample, which included protozoa and yeast-like cells. The entire microbial community was shown to mineralize 75% of the degraded PET into carbon dioxide once it had been initially degraded and assimilated by ''Ideonella sakaiensis''. Characterization Phys ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polyester
Polyester is a category of polymers that contain the ester functional group in every repeat unit of their main chain. As a specific material, it most commonly refers to a type called polyethylene terephthalate (PET). Polyesters include naturally occurring chemicals, such as in plants and insects, as well as synthetics such as polybutyrate. Natural polyesters and a few synthetic ones are biodegradable, but most synthetic polyesters are not. Synthetic polyesters are used extensively in clothing. Polyester fibers are sometimes spun together with natural fibers to produce a cloth with blended properties. Cotton-polyester blends can be strong, wrinkle- and tear-resistant, and reduce shrinking. Synthetic fibers using polyester have high water, wind and environmental resistance compared to plant-derived fibers. They are less Fireproofing, fire-resistant and can melt when ignited. Liquid crystalline polyesters are among the first industrially used liquid crystal polymers. They are use ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PET Bottle Recycling
Although PET is used in several applications, (principally textile fibres for apparel and upholstery, bottles and other rigid packaging, flexible packaging and electrical and electronic goods), as of 2022 only bottles are collected at a substantial scale. The main motivations have been either cost reduction (when oil prices spike) or recycle content of retail goods (driven by regulations or public opinion). An increasing amount is recycled back into bottles, the rest goes into fibres, film, thermoformed packaging and strapping. After sorting, cleaning and grinding, 'bottle flake' is obtained, which is then processed by either: * 'basic' or 'physical' recycling. Bottle flake is melted into its new shape directly with basic changes in its physical properties. * 'chemical' or 'advanced' recycle. Bottle flake (or possibly a less pure feedstock) is partially or totally depolymerized then enabling purification. The resulting oligomers or monomers are repolymerized to PET polymer, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
2-Hydroxyethyl Terephthalic Acid
2-Hydroxyethyl terephthalic acid is an organic compound with the formula HOC2H4O2CC6H4CO2H. It is the monoester of terephthalic acid and ethylene glycol. The compound is a precursor to poly(ethylene terephthalate) (PET), a polymer that is produced on a large scale industrially. 2-Hydroxyethyl terephthalic acid is a colorless solid that is soluble in water and polar organic solvents. Near neutral pH, 2-hydroxyethyl terephthalic acid converts to 2-hydroxyethyl terephthalate, HOC2H4O2CC6H4CO2−. Occurrence and reactions 2-Hydroxyethyl terephthalic acid is an intermediate in both the formation and hydrolysis of PET. It is produced on a massive scale as the first intermediate in certain routes to PET. Specifically, it is produced in the course of the thermal condensation of terephthalic acid and ethylene glycol: :HOC2H4OH + HO2CC6H4CO2H → HOC2H4O2CC6H4CO2H + H2O Further dehydration of 2-hydroxyethyl terephthalic acid gives PET. It is also produced by the partial hydrolys ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Arginine
Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) and both the amino and guanidino groups are protonated, resulting in a cation. Only the -arginine (symbol Arg or R) enantiomer is found naturally. Arg residues are common components of proteins. It is encoded by the codons CGU, CGC, CGA, CGG, AGA, and AGG. The guanidine group in arginine is the precursor for the biosynthesis of nitric oxide. Like all amino acids, it is a white, water-soluble solid. History Arginine was first isolated in 1886 from yellow lupin seedlings by the German chemist Ernst Schulze and his assistant Ernst Steiger. He named it from the Greek ''árgyros'' (ἄργυρος) meaning "silver" due to the silver-white appearance of arginine nitrate crystals. In 1897, Schulze and Ernst Winterstein (1865–1949) determined the structure ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha/beta Hydrolase Superfamily
The alpha/beta hydrolase superfamily is a superfamily of hydrolytic enzymes of widely differing phylogenetic origin and catalytic function that share a common fold. The core of each enzyme is an alpha/beta-sheet (rather than a barrel), containing 8 beta strands connected by 6 alpha helices. The enzymes are believed to have diverged from a common ancestor, retaining little obvious sequence similarity, but preserving the arrangement of the catalytic residues. All have a catalytic triad, the elements of which are borne on loops, which are the best-conserved structural features of the fold. The alpha/beta hydrolase fold includes proteases, lipases, peroxidases, esterases, epoxide hydrolases and dehalogenases. Database The ESTHER database provides a large collection of information about this superfamily of proteins. Subfamilies * 3-oxoadipate enol-lactonase Human proteins containing this domain ABHD10; ABHD11; ABHD12; ABHD12B; ABHD13; ABHD2; ABHD3; ABHD4; ABHD5; ABHD6; A ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pfam
Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models. The most recent version, Pfam 35.0, was released in November 2021 and contains 19,632 families. Uses The general purpose of the Pfam database is to provide a complete and accurate classification of protein families and domains. Originally, the rationale behind creating the database was to have a semi-automated method of curating information on known protein families to improve the efficiency of annotating genomes. The Pfam classification of protein families has been widely adopted by biologists because of its wide coverage of proteins and sensible naming conventions. It is used by experimental biologists researching specific proteins, by structural biologists to identify new targets for structure determination, by computational biologists to organise sequences and by evolutionary biologists tracing the origins of proteins. Early genome ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mutant
In biology, and especially in genetics, a mutant is an organism or a new genetic character arising or resulting from an instance of mutation, which is generally an alteration of the DNA sequence of the genome or chromosome of an organism. It is a characteristic that would not be observed naturally in a specimen. The term mutant is also applied to a virus with an alteration in its nucleotide sequence whose genome is in the nuclear genome. The natural occurrence of genetic mutations is integral to the process of evolution. The study of mutants is an integral part of biology; by understanding the effect that a mutation in a gene has, it is possible to establish the normal function of that gene. Mutants arise by mutation Mutants arise by mutations occurring in pre-existing genomes as a result of errors of DNA replication or errors of DNA repair. Errors of replication often involve translesion synthesis by a DNA polymerase when it encounters and bypasses a damaged base in the temp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L-stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure was estab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycine
Glycine (symbol Gly or G; ) is an amino acid that has a single hydrogen atom as its side chain. It is the simplest stable amino acid (carbamic acid is unstable), with the chemical formula NH2‐ CH2‐ COOH. Glycine is one of the proteinogenic amino acids. It is encoded by all the codons starting with GG (GGU, GGC, GGA, GGG). Glycine is integral to the formation of alpha-helices in secondary protein structure due to its compact form. For the same reason, it is the most abundant amino acid in collagen triple-helices. Glycine is also an inhibitory neurotransmitter – interference with its release within the spinal cord (such as during a ''Clostridium tetani'' infection) can cause spastic paralysis due to uninhibited muscle contraction. It is the only achiral proteinogenic amino acid. It can fit into hydrophilic or hydrophobic environments, due to its minimal side chain of only one hydrogen atom. History and etymology Glycine was discovered in 1820 by the French chemist He ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chymotrypsin
Chymotrypsin (, chymotrypsins A and B, alpha-chymar ophth, avazyme, chymar, chymotest, enzeon, quimar, quimotrase, alpha-chymar, alpha-chymotrypsin A, alpha-chymotrypsin) is a digestive enzyme component of pancreatic juice acting in the duodenum, where it performs proteolysis, the breakdown of proteins and polypeptides. Chymotrypsin preferentially cleaves peptide amide bonds where the side chain of the amino acid N-terminal to the scissile amide bond (the P1 position) is a large hydrophobic amino acid (tyrosine, tryptophan, and phenylalanine). These amino acids contain an aromatic ring in their side chain that fits into a hydrophobic pocket (the S1 position) of the enzyme. It is activated in the presence of trypsin. The hydrophobic and shape complementarity between the peptide substrate P1 side chain and the enzyme S1 binding cavity accounts for the substrate specificity of this enzyme. Chymotrypsin also hydrolyzes other amide bonds in peptides at slower rates, particularly tho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
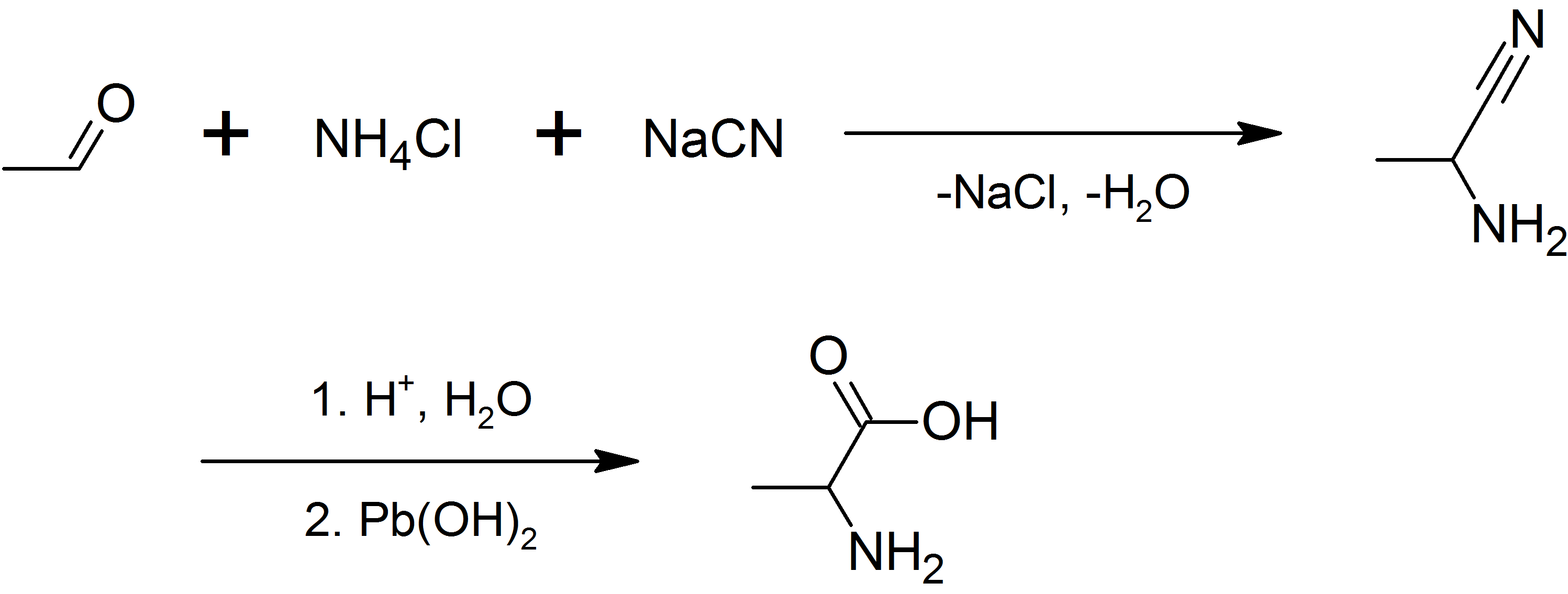
Alanine
Alanine (symbol Ala or A), or α-alanine, is an α-amino acid that is used in the biosynthesis of proteins. It contains an amine group and a carboxylic acid group, both attached to the central carbon atom which also carries a methyl group side chain. Consequently, its IUPAC systematic name is 2-aminopropanoic acid, and it is classified as a nonpolar, aliphatic α-amino acid. Under biological conditions, it exists in its zwitterionic form with its amine group protonated (as −NH3+) and its carboxyl group deprotonated (as −CO2−). It is non-essential to humans as it can be synthesised metabolically and does not need to be present in the diet. It is encoded by all codons starting with GC (GCU, GCC, GCA, and GCG). The L-isomer of alanine (left-handed) is the one that is incorporated into proteins. L-alanine is second only to leucine in rate of occurrence, accounting for 7.8% of the primary structure in a sample of 1,150 proteins. The right-handed form, D-alanine, occurs in p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |