|
Pseudomonas SRNA
''Pseudomonas'' sRNA are non-coding RNAs (ncRNA) that were predicted by the bioinformatic program SRNApredict2. This program identifies putative sRNAs by searching for co-localization of genetic features commonly associated with sRNA-encoding genes and the expression of the predicted sRNAs was subsequently confirmed by Northern blot analysis. These sRNAs have been shown to be conserved across several ''pseudomonas'' species but their function is yet to be determined. Using Tet-Trap genetic approach RNAT genes post-transcriptionally regulated by temperature upshift were identified: ''ptxS'' (implicated in virulence) and PA5194. See also * ''Bacillus subtilis'' sRNA * ''Caenorhabditis elegans'' sRNA * ''Mycobacterium tuberculosis'' sRNA * ''Bacteroides thetaiotaomicron'' sRNA *NrsZ small RNA *AsponA antisense RNA *Repression of heat shock gene expression (ROSE) element *SrbA sRNA SrbA ( sRNA regulator of biofilms A) is a small regulatory non-coding RNA identified in pathogenic ''Ps ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR. The number of non-coding RNAs within the human genome is unknown; however, recent transcriptomic and bioinformatic studies suggest that there are thousands of non-coding transcripts. Many of the newly identified ncRNAs have not been validated for their function. There is no consensus in the literature on how much of non-coding transcription is functional. Some researchers have argued that many ncRNAs are non-functional (sometimes referred to as "junk RNA"), spurious transcriptions. Others, however, disagree, arguing instead that many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for '' in silico'' analyses of biological queries using computational and statistical techniques. Bioinformatics includes biological studies that use computer programming as part of their methodology, as well as specific analysis "pipelines" that are repeatedly used, particularly in the field of genomics. Common uses of bioinformatics include the identification of candidates genes and single nucleotide polymorphisms (SNPs). Often, such identification is made with the aim to better understand the genetic basis of disease, unique adaptations, desirable properties (e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
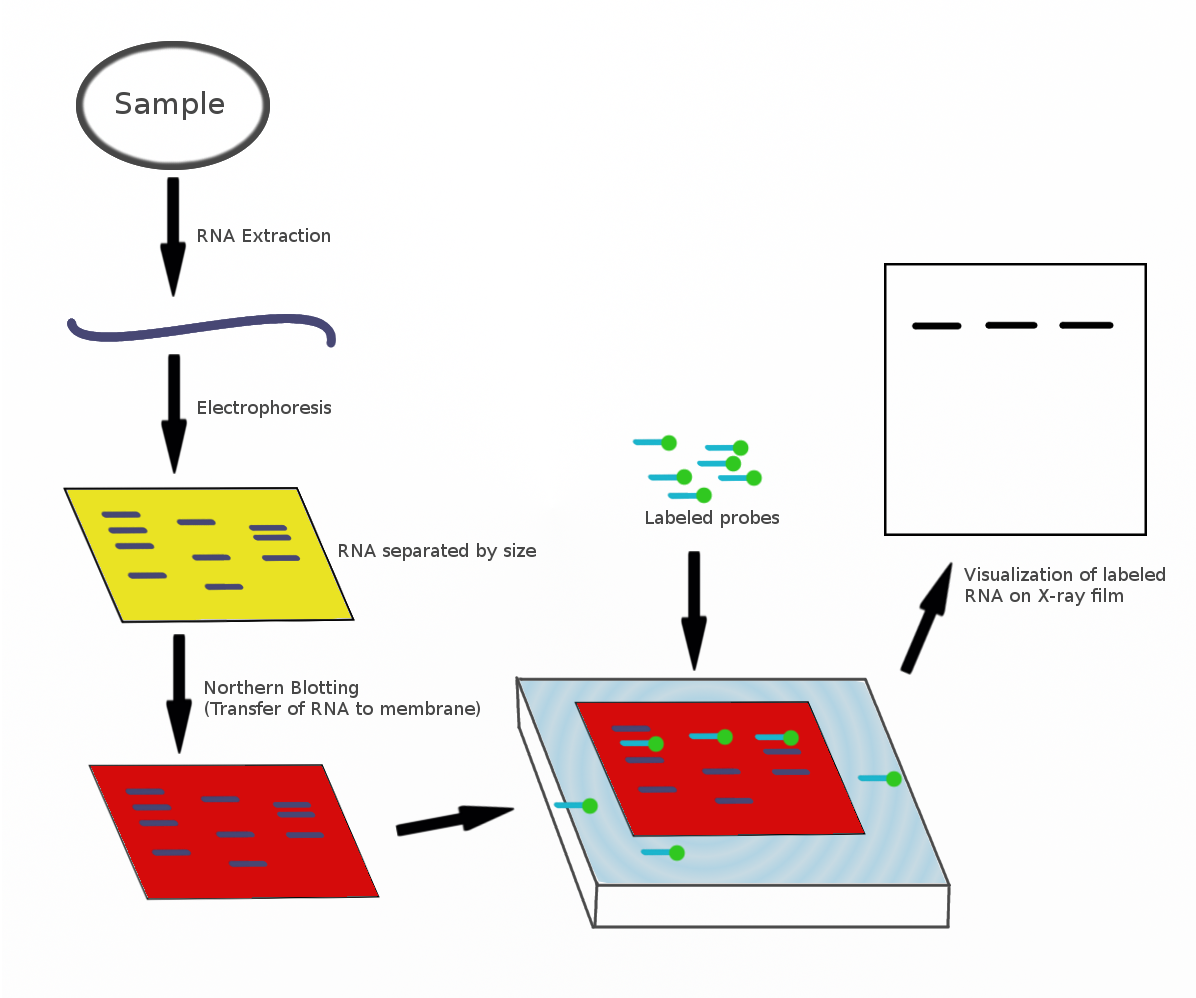
Northern Blot
The northern blot, or RNA blot,Gilbert, S. F. (2000) Developmental Biology, 6th Ed. Sunderland MA, Sinauer Associates. is a technique used in molecular biology research to study gene expression by detection of RNA (or isolated mRNA) in a sample.Kevil, C. G., Walsh, L., Laroux, F. S., Kalogeris, T., Grisham, M. B., Alexander, J. S. (1997) An Improved, Rapid Northern Protocol. Biochem. and Biophys. Research Comm. 238:277–279. With northern blotting it is possible to observe cellular control over structure and function by determining the particular gene expression rates during differentiation and morphogenesis, as well as in abnormal or diseased conditions. Northern blotting involves the use of electrophoresis to separate RNA samples by size, and detection with a hybridization probe complementary to part of or the entire target sequence. Strictly speaking, the term 'northern blot' refers specifically to the capillary transfer of RNA from the electrophoresis gel to the blotting m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conservation (genetics)
In evolutionary biology, conserved sequences are identical or similar Sequence (biology), sequences in nucleic acids (DNA sequence, DNA and RNA) or peptide sequence, proteins across species (homology (biology)#Orthology, orthologous sequences), or within a genome (homology (biology)#Paralogy, paralogous sequences), or between donor and receptor taxa (Sequence homology#Xenology, xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the Ribosomal RNA, RNA components of ribosomes present in all domain (biology), domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudomonas
''Pseudomonas'' is a genus of Gram-negative, Gammaproteobacteria, belonging to the family Pseudomonadaceae and containing 191 described species. The members of the genus demonstrate a great deal of metabolic diversity and consequently are able to colonize a wide range of niches. Their ease of culture ''in vitro'' and availability of an increasing number of ''Pseudomonas'' strain genome sequences has made the genus an excellent focus for scientific research; the best studied species include ''P. aeruginosa'' in its role as an opportunistic human pathogen, the plant pathogen '' P. syringae'', the soil bacterium '' P. putida'', and the plant growth-promoting ''P. fluorescens, P. lini, P. migulae'', and ''P. graminis''. Because of their widespread occurrence in water and plant seeds such as dicots, the pseudomonads were observed early in the history of microbiology. The generic name ''Pseudomonas'' created for these organisms was defined in rather vague terms by Walter Migula i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Thermometer
An RNA thermometer (or RNA thermosensor) is a temperature-sensitive non-coding RNA molecule which regulates gene expression. RNA thermometers often regulate genes required during either a heat shock or cold shock response, but have been implicated in other regulatory roles such as in pathogenicity and starvation. In general, RNA thermometers operate by changing their secondary structure in response to temperature fluctuations. This structural transition can then expose or occlude important regions of RNA such as a ribosome binding site, which then affects the translation rate of a nearby protein-coding gene. RNA thermometers, along with riboswitches, are used as examples in support of the RNA world hypothesis. This theory proposes that RNA was once the sole nucleic acid present in cells, and was replaced by the current DNA → RNA → protein system. Examples of RNA thermometers include FourU, the Hsp90 ''cis''-regulatory element, the ROSE element, the Lig RNA thermom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacillus Subtilis BSR SRNAs
In a screen of the ''Bacillus subtilis'' genome for genes encoding ncRNAs, Saito et al. focused on 123 intergenic regions (IGRs) over 500 base pairs in length, the authors analyzed expression from these regions. Seven IGRs termed bsrC, bsrD, bsrE, bsrF, bsrG, bsrH and bsrI expressed RNAs smaller than 380 nt. All the small RNAs except BsrD RNA were expressed in transformed ''Escherichia coli'' cells harboring a plasmid with PCR-amplified IGRs of ''B. subtilis'', indicating that their own promoters independently express small RNAs. Under non-stressed condition, depletion of the genes for the small RNAs did not affect growth. Although their functions are unknown, gene expression profiles at several time points showed that most of the genes except for bsrD were expressed during the vegetative phase (4–6 h), but undetectable during the stationary phase (8 h). Mapping the 5' ends of the 6 small RNAs revealed that the genes for BsrE, BsrF, BsrG, BsrH, and BsrI RNAs are preceded by a rec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mycobacterium Tuberculosis SRNA
''Mycobacterium tuberculosis'' contains at least nine small RNA families in its genome. The small RNA (sRNA) families were identified through RNomics – the direct analysis of RNA molecules isolated from cultures of ''Mycobacterium tuberculosis''. The sRNAs were characterised through RACE mapping and Northern blot experiments. Secondary structures of the sRNAs were predicted using Mfold. sRNAPredict2 – a bioinformatics tool – suggested 56 putative sRNAs in ''M. tuberculosis'', though these have yet to be verified experimentally. Hfq protein homologues have yet to be found in ''M. tuberculosis''; an alternative pathway – potentially involving conserved C-rich motifs – has been theorised to enable trans-acting sRNA functionality. sRNAs were shown to have important physiological roles in ''M. tuberculosis''. Overexpression of G2 sRNA, for example, prevented growth of ''M. tuberculosis'' and greatly reduced the growth of ''M. smegmatis''; ASdes sRNA is thought to be a cis-a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteroides Thetaiotaomicron SRNA
The ''Bacteroides thetaiotaomicron'' genome contains hundreds of small RNAs (sRNAs), discovered through RNA sequencing. These include canonical housekeeping RNA species such as the 6S RNA (SsrS), tmRNA (SsrA), M1 RNA (RnpB) and 4.5S RNA (Ffs) as well as several hundred cis and trans encoded small RNAs. More than 20 candidates have been validated with northern blots and the structures of several members have been characterized through ''in silico'' analyses and chemical probing experiments. Two ''B. thetaiotaomicron'' sRNAs that have been functionally characterized are RteR and GibS. RteR is a 78 nucleotide (nt) long sRNA that is conserved in closely related species and likely serves as a repressor of a transposon operon. Analyses based on secondary structure conservation, taking into consideration nucleotide covariation and in-vitro chemical probing have revealed a structure that consists of a 5’ hairpin and a Rho-independent terminator that are separated by an 8 nt sequ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NrsZ Small RNA
NrsZ (nitrogen regulated small RNA) is a bacterial small RNA found in the opportunistic pathogen ''Pseudomonas aeruginosa'' PAO1. Its transcription is induced during nitrogen limitation by the NtrB/C two-component system (an important regulator of nitrogen assimilation and swarming motility) together with the alternative sigma factor RpoN ( a global regulator involved in nitrogen metabolism). NrsZ by activating ''rhlA (''a gene essential for rhamnolipids synthesis) positively regulates the production of rhamnolipid surfactants needed for swarming motility. See also * ''Pseudomonas'' sRNA * SrbA sRNA * AsponA antisense RNA AsponA is a small asRNA transcribed antisense to the penicillin-binding protein 1A gene called ''ponA.'' It was identified by RNAseq and the expression was validated by 5' and 3' RACE experiments in '' Pseudomanas aeruginosa''. ''AsponA'' express ... References Non-coding RNA {{molecular-cell-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
AsponA Antisense RNA
AsponA is a small asRNA transcribed antisense to the penicillin-binding protein 1A gene called ''ponA.'' It was identified by RNAseq and the expression was validated by 5' and 3' RACE experiments in '' Pseudomanas aeruginosa''. ''AsponA'' expression was up or down regulated under different antibiotic stress. Owing to its location it may be able to prevent the transcription or translation of the opposite gene. Study by Wurtzel ''et al.'' and Ferrara ''et al.'' also detected its expression. See also * NrsZ small RNA * SrbA sRNA SrbA ( sRNA regulator of biofilms A) is a small regulatory non-coding RNA identified in pathogenic ''Pseudomonas aeruginosa'.'' It is important for biofilm formation and pathogenicity. Bacterial strain with deleted SrbA had reduced biofilm mass. A ... * ''Pseudomonas'' sRNA References {{Reflist Non-coding RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |