|
Positive-sense Single-stranded RNA Virus
Positive-strand RNA viruses (+ssRNA viruses) are a group of related viruses that have positive-sense, single-stranded genomes made of ribonucleic acid. The positive-sense genome can act as messenger RNA (mRNA) and can be directly translated into viral proteins by the host cell's ribosomes. Positive-strand RNA viruses encode an RNA-dependent RNA polymerase (RdRp) which is used during replication of the genome to synthesize a negative-sense antigenome that is then used as a template to create a new positive-sense viral genome. Positive-strand RNA viruses are divided between the phyla '' Kitrinoviricota'', '' Lenarviricota'', and '' Pisuviricota'' (specifically classes '' Pisoniviricetes'' and '' Stelpavirictes'') all of which are in the kingdom '' Orthornavirae'' and realm ''Riboviria''. They are monophyletic and descended from a common RNA virus ancestor. In the Baltimore classification system, +ssRNA viruses belong to Group IV. Positive-sense RNA viruses include patho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
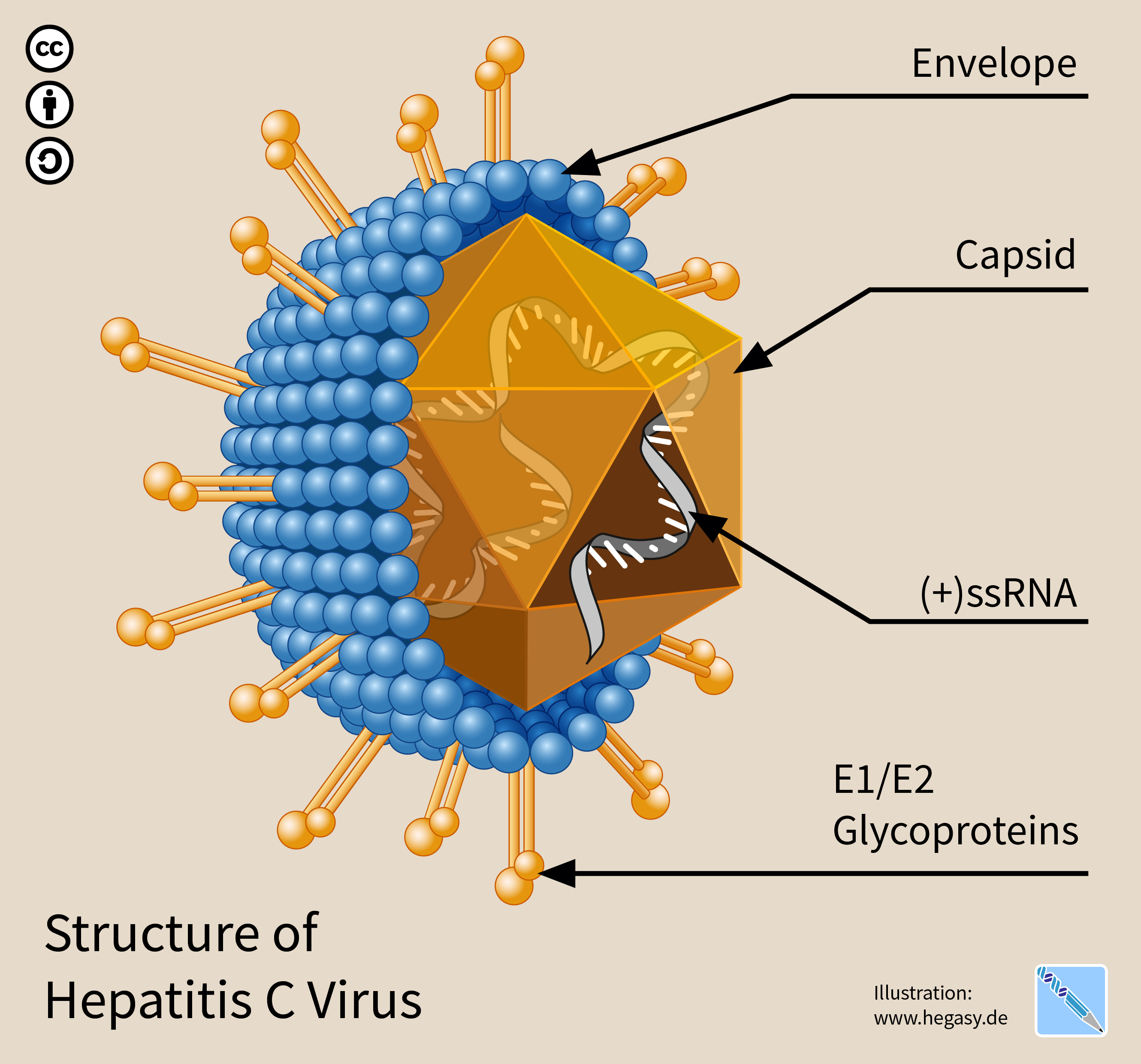
Hepatitis C Virus
The hepatitis C virus (HCV) is a small (55–65 nm in size), enveloped, positive-sense single-stranded RNA virus of the family ''Flaviviridae''. The hepatitis C virus is the cause of hepatitis C and some cancers such as liver cancer ( hepatocellular carcinoma, abbreviated HCC) and lymphomas in humans. Taxonomy The hepatitis C virus belongs to the genus '' Hepacivirus'', a member of the family ''Flaviviridae''. Before 2011, it was considered to be the only member of this genus. However a member of this genus has been discovered in dogs: canine hepacivirus. There is also at least one virus in this genus that infects horses. Several additional viruses in the genus have been described in bats and rodents. Structure The hepatitis C virus particle consists of a lipid membrane envelope that is 55 to 65 nm in diameter. Two viral envelope glycoproteins, E1 and E2, are embedded in the lipid envelope. They take part in viral attachment and entry into the cell. Within the e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monophyly
In biological cladistics for the classification of organisms, monophyly is the condition of a taxonomic grouping being a clade – that is, a grouping of organisms which meets these criteria: # the grouping contains its own most recent common ancestor (or more precisely an ancestral population), i.e. excludes non-descendants of that common ancestor # the grouping contains all the descendants of that common ancestor, without exception Monophyly is contrasted with paraphyly and polyphyly as shown in the second diagram. A ''paraphyletic'' grouping meets 1. but not 2., thus consisting of the descendants of a common ancestor, excepting one or more monophyletic subgroups. A ''polyphyletic'' grouping meets neither criterion, and instead serves to characterize convergent relationships of biological features rather than genetic relationships – for example, night-active primates, fruit trees, or aquatic insects. As such, these characteristic features of a polyphyletic grouping are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exoribonuclease
An exoribonuclease is an exonuclease ribonuclease, which are enzymes that degrade RNA by removing terminal nucleotides from either the 5' end or the 3' end of the RNA molecule. Enzymes that remove nucleotides from the 5' end are called ''5'-3' exoribonucleases'', and enzymes that remove nucleotides from the 3' end are called ''3'-5' exoribonucleases''. Exoribonucleases can use either water to cleave the nucleotide-nucleotide bond (which is called hydrolytic activity) or inorganic phosphate (which is called phosphorolytic activity). Hydrolytic exoribonucleases are classified under EC number 3.1 and phosphorolytic exoribonucleases under EC number 2.7.7. As the phosphorolytic enzymes use inorganic phosphate to cleave bonds they release nucleotide diphosphates, whereas the hydrolytic enzymes (which use water) release nucleotide monosphosphates. Exoribonucleases exist in all kingdoms of life, the bacteria, archaea, and eukaryotes. Exoribonucleases are involved in the degradati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Kilobase
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine– thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Common Cold
The common cold, or the cold, is a virus, viral infectious disease of the upper respiratory tract that primarily affects the Respiratory epithelium, respiratory mucosa of the human nose, nose, throat, Paranasal sinuses, sinuses, and larynx. Signs and symptoms may appear in as little as two days after exposure to the virus. These may include coughing, sore throat, rhinorrhea, runny nose, Sneeze, sneezing, headache, fatigue, and fever. People usually recover in seven to ten days, but some symptoms may last up to three weeks. Occasionally, those with other health problems may develop pneumonia. Well over 200 virus strains are implicated in causing the common cold, with rhinoviruses, coronaviruses, Adenoviridae, adenoviruses and enteroviruses being the most common. They spread through the air or indirectly through contact with objects in the environment, followed by transfer to the mouth or nose. Risk factors include going to child care facilities, Sleep deprivation, not sleepin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rhinovirus
The rhinovirus (from the "nose", , romanized: "of the nose", and the ) is a Positive-sense single stranded RNA virus, positive-sense, single-stranded RNA virus belonging to the genus ''Enterovirus'' in the family ''Picornaviridae''. Rhinovirus is the most common viral infectious agent in humans and is the predominant cause of the common cold. The three species of rhinovirus (A, B, and C) include at least 165 recognized types that differ according to their surface antigens or genetics. They are among the smallest viruses, with diameters of about 30 nanometers. By comparison, other viruses, such as smallpox and vaccinia, are around ten times larger at about 300 nanometers, while Orthomyxoviridae, influenza viruses are around 80–120 nm. Rhinoviruses are transmitted through aerosols, respiratory droplets, fomites, and direct person-to-person contact. They primarily infect nasal epithelial cells in the airway and cause mild symptoms such as sore throat, cough, and nasal con ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coronavirus
Coronaviruses are a group of related RNA viruses that cause diseases in mammals and birds. In humans and birds, they cause respiratory tract infections that can range from mild to lethal. Mild illnesses in humans include some cases of the common cold (which is also caused by other viruses, predominantly rhinoviruses), while more lethal varieties can cause SARS, MERS and COVID-19. In cows and pigs they cause diarrhea, while in mice they cause hepatitis and encephalomyelitis. Coronaviruses constitute the subfamily ''Orthocoronavirinae'', in the family ''Coronaviridae'', order ''Nidovirales'' and realm ''Riboviria''. They are enveloped viruses with a Positive-strand RNA virus, positive-sense single-stranded RNA genome and a nucleocapsid of helical symmetry. The genome size of coronaviruses ranges from approximately 26 to 32 kilobases, one of the largest among RNA viruses. They have characteristic club-shaped Spike protein, spikes that project from their surface, which in electron ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) is a strain of coronavirus that causes COVID-19, the respiratory illness responsible for the COVID-19 pandemic. The virus previously had the Novel coronavirus, provisional name 2019 novel coronavirus (2019-nCoV), and has also been called human coronavirus 2019 (HCoV-19 or hCoV-19). First identified in the city of Wuhan, Hubei, China, the World Health Organization designated the outbreak a public health emergency of international concern from January 30, 2020, to May 5, 2023. SARS‑CoV‑2 is a positive-sense single-stranded RNA virus that is Contagious disease, contagious in humans. SARS‑CoV‑2 is a strain of the species ''Betacoronavirus pandemicum'' (SARSr-CoV), as is SARS-CoV-1, the virus that caused the 2002–2004 SARS outbreak. There are animal-borne coronavirus strains more closely related to SARS-CoV-2, the most closely known relative being the BANAL-52 bat coronavirus. SARS-CoV-2 is of Zoonosis, z ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SARS Coronavirus
Severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1), previously known as severe acute respiratory syndrome coronavirus (SARS-CoV), is a strain of coronavirus that causes severe acute respiratory syndrome (SARS), the respiratory illness responsible for the 2002–2004 SARS outbreak. It is an enveloped, positive-sense, single-stranded RNA virus that infects the epithelial cells within the lungs. The virus enters the host cell by binding to angiotensin-converting enzyme 2. It infects humans, bats, and palm civets. The SARS-CoV-1 outbreak was largely brought under control by simple public health measures. Testing people with symptoms (fever and respiratory problems), isolating and quarantining suspected cases, and restricting travel all had an effect. SARS-CoV-1 was most transmissible when patients were sick, so its spread could be effectively suppressed by isolating patients with symptoms. On April 16, 2003, following the outbreak of SARS in Asia and secondary c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MERS Coronavirus
Middle East respiratory syndrome–related coronavirus (MERS-CoV, ''Betacoronavirus cameli'') or EMC/2012 ( HCoV-EMC/2012), is the virus that causes Middle East respiratory syndrome (MERS). It is a species of coronavirus which infects humans, bats, and camels. The infecting virus is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the DPP4 receptor. The species is a member of the genus ''Betacoronavirus'' and subgenus '' Merbecovirus''. Initially called simply novel coronavirus or nCoV, with the provisional names 2012 novel coronavirus (2012-nCoV) and human coronavirus 2012 (HCoV-12 or hCoV-12), it was first reported in June 2012 after genome sequencing of a virus isolated from sputum samples from a person who fell ill in a 2012 outbreak of a new flu-like respiratory illness. By July 2015, MERS-CoV cases had been reported in over 21 countries, in Europe, North America and Asia as well as the Middle East. MERS-CoV is one of seve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
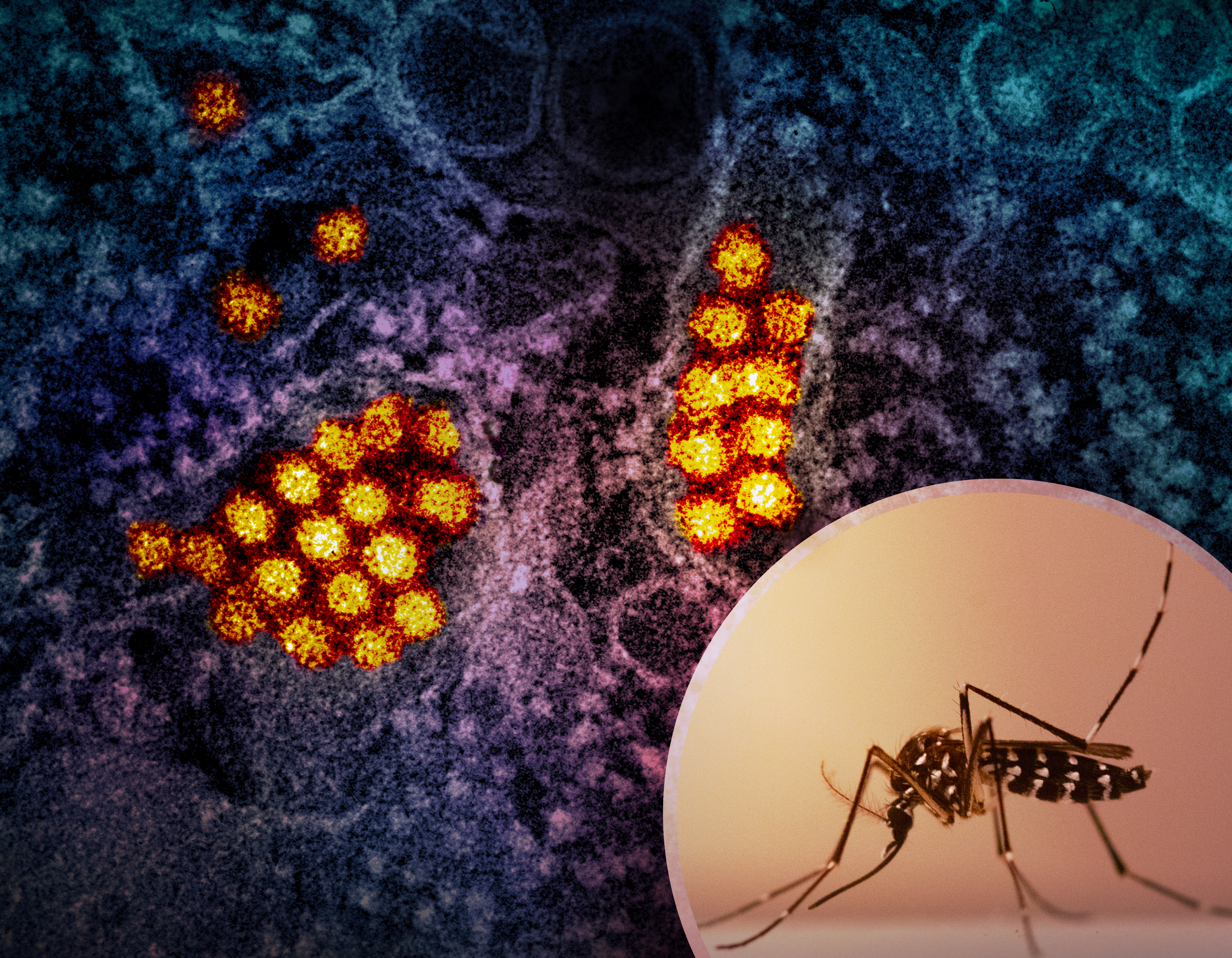
Dengue Virus
Dengue virus (DENV) is the cause of dengue fever. It is a mosquito-borne, single positive-stranded RNA virus of the family ''Flaviviridae''; genus '' Flavivirus''. Four serotypes of the virus have been found, and a reported fifth has yet to be confirmed,Dwivedi, V. D., Tripathi, I. P., Tripathi, R. C., Bharadwaj, S., & Mishra, S. K. (2017). Genomics, proteomics and evolution of ''Dengue virus''. Briefings in functional genomics.16(4): 217–227, https://doi.org/10.1093/bfgp/elw040 all of which can cause the full spectrum of disease. Nevertheless, the mainstream scientific community's understanding of dengue virus may be simplistic as, rather than distinct antigenic groups, a continuum appears to exist. This same study identified 47 strains of ''dengue virus''. Additionally, coinfection with and lack of rapid tests for Zika virus and chikungunya complicate matters in real-world infections. ''Dengue virus'' has increased dramatically within the last 20 years, becoming one of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |