|
ORF3b
ORF3b is a gene found in coronaviruses of the subgenus ''Sarbecovirus'', encoding a short non-structural protein. It is present in both SARS-CoV (which causes the disease SARS) and SARS-CoV-2 (which causes COVID-19), though the protein product has very different lengths in the two viruses. The encoded protein is significantly shorter in SARS-CoV-2, at only 22 amino acid residues compared to 153-155 in SARS-CoV. Both the longer SARS-CoV and shorter SARS-CoV-2 proteins have been reported as interferon antagonists. It is unclear whether the SARS-CoV-2 gene expresses a functional protein. Nomenclature There has been significant confusion in the scientific literature around the nomenclature used for the accessory proteins of SARS-CoV-2, especially several overlapping genes with ORF3a. Due to differences in the genomes of SARS-CoV and SARS-CoV-2, two distinct open reading frames (ORFs) in the SARS-CoV-2 genome have been referred to as "ORF3b". In SARS-CoV, ORF3b is a gene of 155 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ORF3d
ORF3d is a gene found in SARS-CoV-2 (the virus that causes COVID-19) and at least one closely related coronavirus found in pangolins, though it is not found in other closely related viruses within the ''Sarbecovirus'' subgenus. It is 57 codons long and encodes a novel 57 amino acid residue protein of unknown function. At least two isoforms have been described, of which the shorter 33-residue form, ORF3d-2, may be more highly expressed, or even the only form expressed. It is reported to be antigenic and antibodies to the ORF3d protein occur in patients recovered from COVID-19. There is no homolog in the genome of the otherwise closely related SARS-CoV (which causes the disease SARS). Nomenclature There has been significant confusion in the scientific literature around the nomenclature used for the accessory proteins of SARS-CoV-2, especially several overlapping genes with ORF3a. Many scientific papers have referred to ORF3d and its protein product as ORF3b, due to confusion ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ORF3a
ORF3a (previously known as X1 or U274) is a gene found in coronaviruses of the subgenus ''Sarbecovirus'', including SARS-CoV and SARS-CoV-2. It encodes an accessory protein about 275 amino acid residues long, which is thought to function as a viroporin. It is the largest accessory protein and was the first of the SARS-CoV accessory proteins to be described. Comparative genomics ORF3a is well conserved within the subgenus ''Sarbecovirus''. The protein has 73% sequence identity between SARS-CoV (274 residues) and SARS-CoV-2 (275 residues). Within the ORF3a open reading frame there are several overlapping genes in the genome: ORF3a, ORF3b, and (in SARS-CoV-2 only) ORF3c. In SARS-CoV-2, the overlap between ORF3a, ORF3c, and ORF3d potentially represents a rare example of all three possible reading frames of the same sequence region encoding functional proteins. Although ORF3a is present in ''Sarbecovirus'', it is absent in another ''Betacoronavirus'' subgenus, ''Embecovirus'', which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
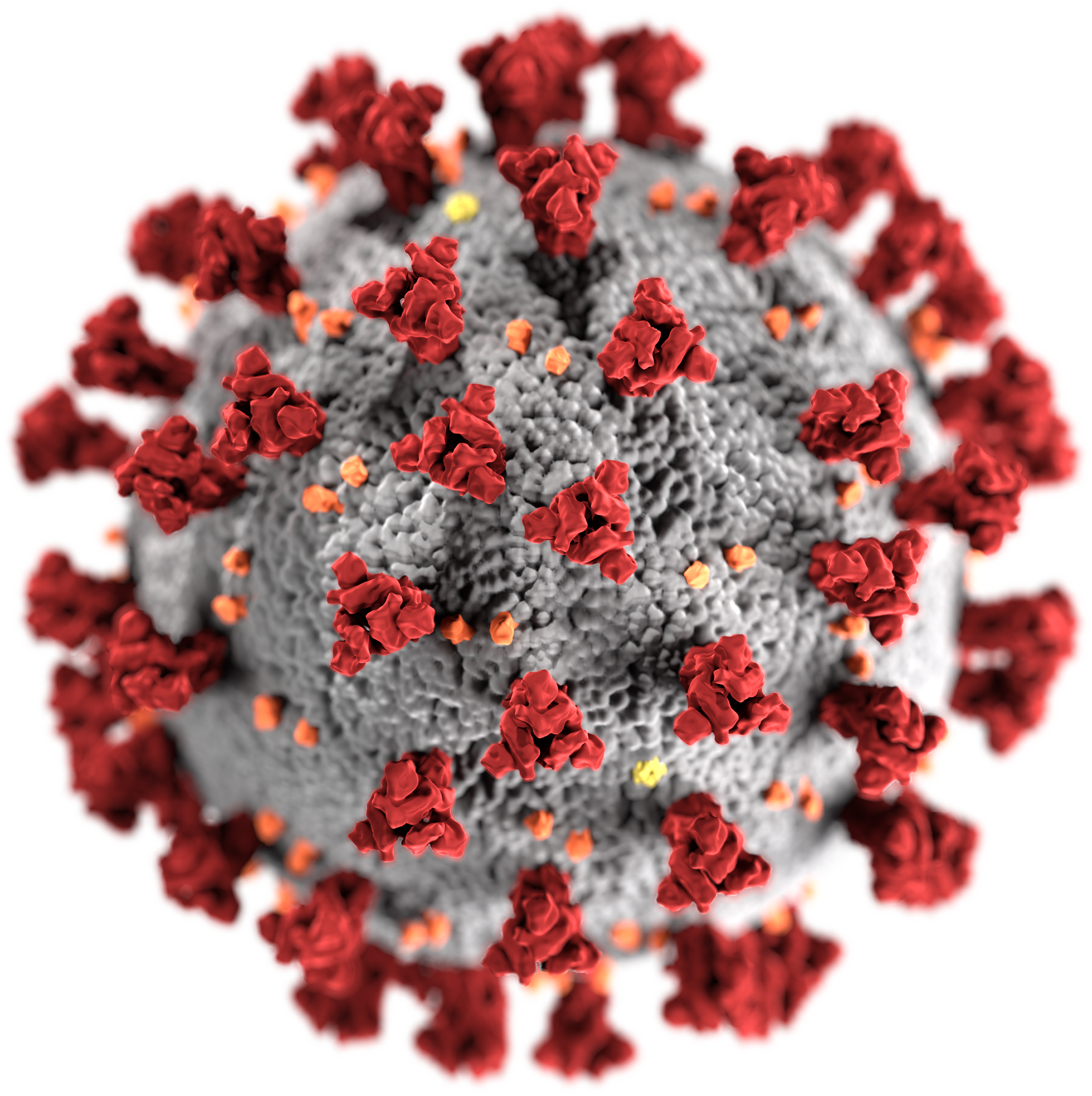
Sarbecovirus
''Severe acute respiratory syndrome–related coronavirus'' (SARSr-CoV or SARS-CoV)The terms ''SARSr-CoV'' and ''SARS-CoV'' are sometimes used interchangeably, especially prior to the discovery of SARS-CoV-2. This may cause confusion when some publications refer to SARS-CoV-1 as ''SARS-CoV''. is a species of virus consisting of many known strains phylogenetically related to severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) that have been shown to possess the capability to infect humans, bats, and certain other mammals. These enveloped, positive-sense single-stranded RNA viruses enter host cells by binding to the angiotensin-converting enzyme 2 (ACE2) receptor. The SARSr-CoV species is a member of the genus ''Betacoronavirus'' and of the subgenus ''Sarbecovirus'' (SARS Betacoronavirus). Two strains of the virus have caused outbreaks of severe respiratory diseases in humans: severe acute respiratory syndrome coronavirus 1 (SARS-CoV or SARS-CoV-1), which caused the 20 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SARSr-CoV
''Severe acute respiratory syndrome–related coronavirus'' (SARSr-CoV or SARS-CoV)The terms ''SARSr-CoV'' and ''SARS-CoV'' are sometimes used interchangeably, especially prior to the discovery of SARS-CoV-2. This may cause confusion when some publications refer to SARS-CoV-1 as ''SARS-CoV''. is a species of virus consisting of many known strains phylogenetically related to severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) that have been shown to possess the capability to infect humans, bats, and certain other mammals. These enveloped, positive-sense single-stranded RNA viruses enter host cells by binding to the angiotensin-converting enzyme 2 (ACE2) receptor. The SARSr-CoV species is a member of the genus ''Betacoronavirus'' and of the subgenus ''Sarbecovirus'' (SARS Betacoronavirus). Two strains of the virus have caused outbreaks of severe respiratory diseases in humans: severe acute respiratory syndrome coronavirus 1 (SARS-CoV or SARS-CoV-1), which caused the 20 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ORF3c
ORF3c is a gene found in coronaviruses of the subgenus ''Sarbecovirus'', including SARS-CoV and SARS-CoV-2. It was first identified in the SARS-CoV-2 genome and encodes a 41 amino acid non-structural protein of unknown function. It is also present in the SARS-CoV genome, but was not recognized until the identification of the SARS-CoV-2 homolog. Nomenclature There has been significant confusion in the scientific literature around the nomenclature used for the accessory proteins of SARS-CoV-2, especially several overlapping genes with ORF3a. The predicted protein product of the ''ORF3c'' gene has at least once been referred to as "3b protein", but it is not to be confused with the non-homologous gene '' ORF3b''. It has also been described under the names ''ORF3h'' and ''ORF3a.iORF1''. The recommended nomenclature for SARS-CoV-2 uses the term ''ORF3c'' for this gene. Comparative genomics ORF3c is an overlapping gene whose open reading frame overlaps both ORF3a and ORF3d in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stop Codon
In molecular biology (specifically protein biosynthesis), a stop codon (or termination codon) is a codon (nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain. While start codons need nearby sequences or initiation factors to start translation, a stop codon alone is sufficient to initiate termination. Properties Standard codons In the standard genetic code, there are three different termination codons: Alternative stop codons There are variations on the standard genetic code, and alternative stop codons have been found in the mitochondrial genomes of vertebrates, ''Scenedesmus obliquus'', and '' Thra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a protein often c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Localization Signal
A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different nuclear localized proteins may share the same NLS. An NLS has the opposite function of a nuclear export signal (NES), which targets proteins out of the nucleus. Types Classical These types of NLSs can be further classified as either monopartite or bipartite. The major structural differences between the two are that the two basic amino acid clusters in bipartite NLSs are separated by a relatively short spacer sequence (hence bipartite - 2 parts), while monopartite NLSs are not. The first NLS to be discovered was the sequence PKKKRKV in the SV40 Large T-antigen (a monopartite NLS). The NLS of nucleoplasmin, KR AATKKAGQAKKK, is the prototype of the ubiquitous bipartite signal: two cluster ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Internal Ribosome Entry Site
An internal ribosome entry site, abbreviated IRES, is an RNA element that allows for translation initiation in a cap-independent manner, as part of the greater process of protein synthesis. In eukaryotic translation, initiation typically occurs at the 5' end of mRNA molecules, since 5' cap recognition is required for the assembly of the initiation complex. The location for IRES elements is often in the 5'UTR, but can also occur elsewhere in mRNAs. History IRES sequences were first discovered in 1988 in the poliovirus (PV) and encephalomyocarditis virus (EMCV) RNA genomes in the labs of Nahum Sonenberg and Eckard Wimmer, respectively. They are described as distinct regions of RNA molecules that are able to recruit the eukaryotic ribosome to the mRNA. This process is also known as cap-independent translation. It has been shown that IRES elements have a distinct secondary or even tertiary structure, but similar structural features at the levels of either primary or secondary structur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translation (biology)
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription (biology), transcription of DNA to RNA in the cell's nucleus (cell), nucleus. The entire process is called gene expression. In translation, mRNA, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later protein folding, folds into an Activation energy, active protein and performs its functions in the Cell (biology), cell. The ribosome facilitates decoding by inducing the binding of Base pair, complementary tRNA anticodon sequences to mRNA codons. The tRNAs carry specific amino acids that are chained together into a polypeptide as the mRNA passes through and is "read" by the ribosome. Translation proceeds in three phases: # Initiation: The ribosome assembles around the target mRNA. The first tRNA is attached a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Conservation
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, promp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |