|
Nucleolar
The nucleolus (, plural: nucleoli ) is the largest structure in the nucleus of eukaryotic cells. It is best known as the site of ribosome biogenesis, which is the synthesis of ribosomes. The nucleolus also participates in the formation of signal recognition particles and plays a role in the cell's response to stress. Nucleoli are made of proteins, DNA and RNA, and form around specific chromosomal regions called nucleolar organizing regions. Malfunction of nucleoli can be the cause of several human conditions called "nucleolopathies" and the nucleolus is being investigated as a target for cancer chemotherapy. History The nucleolus was identified by bright-field microscopy during the 1830s. Little was known about the function of the nucleolus until 1964, when a study of nucleoli by John Gurdon and Donald Brown in the African clawed frog ''Xenopus laevis'' generated increasing interest in the function and detailed structure of the nucleolus. They found that 25% of the frog eg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal RNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form SSU rRNA, small and LSU rRNA, large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and Translation (biology), translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass. Structure Although the primary structure of rRNA sequences can vary across organisms, Base pair, base-pairing within these sequences commonly forms stem-loop configurations. The length and position of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
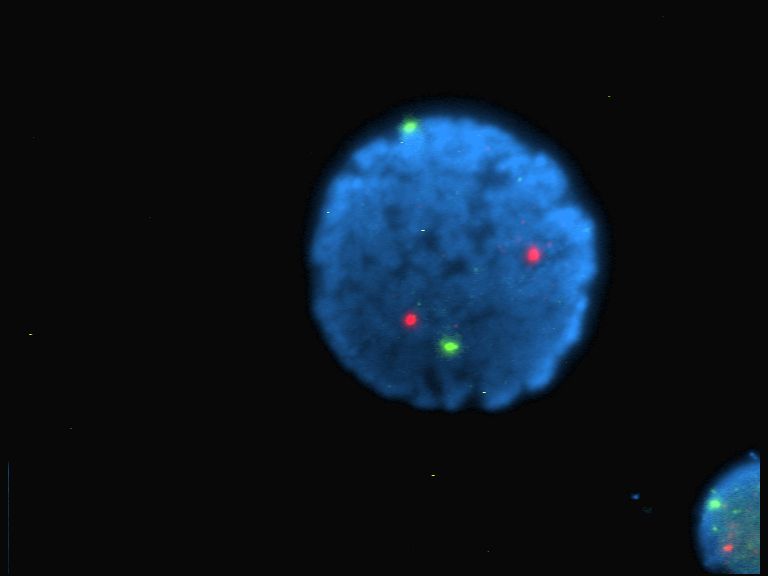
Nucleolus Organizer Region
] Nucleolus organizer regions (NORs) are chromosome, chromosomal regions crucial for the formation of the nucleolus. In humans, the NORs are located on the short arms of the acrocentric chromosomes 13, 14, 15, 21 and 22, the genes RNR1, RNR2, RNR3, RNR4, and RNR5 respectively. These regions code for 5.8S, 18S, and 28S ribosomal RNA. The NORs are "sandwiched" between the repetitive, heterochromatic DNA sequences of the centromeres and telomeres. The exact sequence of these regions is not included in the human reference genome as of 2016 or the GRCh38.p10 released January 6, 2017. On 28 February 2019, GRCh38.p13 was released, which added the NOR sequences for the short arms of chromosomes 13, 14, 15, 21, and 22. However, it is known that NORs contain tandem copies of ribosomal DNA (rDNA) genes. Some sequences of flanking sequences proximal and distal to NORs have been reported. The NORs of a loris have been reported to be highly variable. There are also DNA sequences related to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleolar Organizing Regions
] Nucleolus organizer regions (NORs) are chromosome, chromosomal regions crucial for the formation of the nucleolus. In humans, the NORs are located on the short arms of the acrocentric chromosomes 13, 14, 15, 21 and 22, the genes RNR1, RNR2, RNR3, RNR4, and RNR5 respectively. These regions code for 5.8S, 18S, and 28S ribosomal RNA. The NORs are "sandwiched" between the repetitive, heterochromatic DNA sequences of the centromeres and telomeres. The exact sequence of these regions is not included in the human reference genome as of 2016 or the GRCh38.p10 released January 6, 2017. On 28 February 2019, GRCh38.p13 was released, which added the NOR sequences for the short arms of chromosomes 13, 14, 15, 21, and 22. However, it is known that NORs contain tandem copies of ribosomal DNA (rDNA) genes. Some sequences of flanking sequences proximal and distal to NORs have been reported. The NORs of a loris have been reported to be highly variable. There are also DNA sequences related t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NPM1
Nucleophosmin (NPM), also known as nucleolar phosphoprotein B23 or numatrin, is a protein that in humans is encoded by the ''NPM1'' gene. Function NPM1 is associated with nucleolar ribonucleoprotein structures and binds single-stranded and double-stranded nucleic acids, but it binds preferentially G-quadruplex forming nucleic acids. It is involved in the biogenesis of ribosomes and may assist small basic proteins in their transport to the nucleolus. Its regulation through SUMOylation (by SENP3 and SENP5) is another facet of the protein's regulation and cellular functions. It is located in the nucleolus, but it can be translocated to the nucleoplasm in case of serum starvation or treatment with anticancer drugs. The protein is phosphorylated. Nucleophosmin has multiple functions: # Histone chaperones # Ribosome biogenesis and transport # Genomic stability and DNA repair # Endoribonuclease activity # Centrosome duplication during cell cycle # Regulation of ARF-p53 tumor supp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal DNA
Ribosomal DNA (rDNA) is a DNA sequence that codes for ribosomal RNA. These sequences regulate transcription initiation and amplification, and contain both transcribed and non-transcribed spacer segments. In the human genome there are 5 chromosomes with nucleolus organizer regions: the acrocentric chromosomes 13 (RNR1), 14 ( RNR2), 15 ( RNR3), 21 (RNR4) and 22 (RNR5). The genes that are responsible for encoding the various sub-units of rRNA are located across multiple chromosomes in humans. But the genes that encode for rRNA are highly conserved across the domains, with only the copy numbers involved for the genes having varying numbers per species. In Bacteria, Archaea, and chloroplasts the rRNA is composed of different (smaller) units, the large (23S) ribosomal RNA, 16S ribosomal RNA and 5S rRNA. The 16S rRNA is widely used for phylogenetic studies. Eukaryotes The rRNA transcribed from the approximately 600 rDNA repeats forms the most abundant section of RNA found in cells ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescence Recovery After Photobleaching
Fluorescence recovery after photobleaching (FRAP) is a method for determining the kinetics of diffusion through tissue or cells. It is capable of quantifying the two dimensional lateral diffusion of a molecularly thin film containing fluorescently labeled probes, or to examine single cells. This technique is very useful in biological studies of cell membrane diffusion and protein binding. In addition, surface deposition of a fluorescing phospholipid bilayer (or monolayer) allows the characterization of hydrophilic (or hydrophobic) surfaces in terms of surface structure and free energy. Similar, though less well known, techniques have been developed to investigate the 3-dimensional diffusion and binding of molecules inside the cell; they are also referred to as FRAP. Experimental setup The basic apparatus comprises an optical microscope, a light source and some fluorescent probe. Fluorescent emission is contingent upon absorption of a specific optical wavelength or color which re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Photobleaching
In optics, photobleaching (sometimes termed fading) is the photochemical alteration of a dye or a fluorophore molecule such that it is permanently unable to fluoresce. This is caused by cleaving of covalent bonds or non-specific reactions between the fluorophore and surrounding molecules. Such irreversible modifications in covalent bonds are caused by transition from a singlet state to the triplet state of the fluorophores. The number of excitation cycles to achieve full bleaching varies. In microscopy, photobleaching may complicate the observation of fluorescent molecules, since they will eventually be destroyed by the light exposure necessary to stimulate them into fluorescing. This is especially problematic in time-lapse microscopy. However, photobleaching may also be used prior to applying the (primarily antibody-linked) fluorescent molecules, in an attempt to quench autofluorescence. This can help improve the signal-to-noise ratio. Photobleaching may also be exploited to study ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorophore
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with several π bonds. Fluorophores are sometimes used alone, as a tracer in fluids, as a dye for staining of certain structures, as a substrate of enzymes, or as a probe or indicator (when its fluorescence is affected by environmental aspects such as polarity or ions). More generally they are covalently bonded to a macromolecule, serving as a marker (or dye, or tag, or reporter) for affine or bioactive reagents (antibodies, peptides, nucleic acids). Fluorophores are notably used to stain tissues, cells, or materials in a variety of analytical methods, i.e., fluorescent imaging and spectroscopy. Fluorescein, via its amine-reactive isothiocyanate derivative fluorescein isothiocyanate (FITC), has been one of the most popular fluorophores. Fro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electron Microscope
An electron microscope is a microscope that uses a beam of accelerated electrons as a source of illumination. As the wavelength of an electron can be up to 100,000 times shorter than that of visible light photons, electron microscopes have a higher resolving power than light microscopes and can reveal the structure of smaller objects. A scanning transmission electron microscope has achieved better than 50 pm resolution in annular dark-field imaging mode and magnifications of up to about 10,000,000× whereas most light microscopes are limited by diffraction to about 200 nm resolution and useful magnifications below 2000×. Electron microscopes use shaped magnetic fields to form electron optical lens systems that are analogous to the glass lenses of an optical light microscope. Electron microscopes are used to investigate the ultrastructure of a wide range of biological and inorganic specimens including microorganisms, cells, large molecules, biopsy samples, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ultrastructure
Ultrastructure (or ultra-structure) is the architecture of cells and biomaterials that is visible at higher magnifications than found on a standard optical light microscope. This traditionally meant the resolution and magnification range of a conventional transmission electron microscope (TEM) when viewing biological specimens such as cells, tissue, or organs. Ultrastructure can also be viewed with scanning electron microscopy and super-resolution microscopy, although TEM is a standard histology technique for viewing ultrastructure. Such cellular structures as organelles, which allow the cell to function properly within its specified environment, can be examined at the ultrastructural level. Ultrastructure, along with molecular phylogeny, is a reliable phylogenetic way of classifying organisms. Features of ultrastructure are used industrially to control material properties and promote biocompatibility. History In 1931, German engineers Max Knoll and Ernst Ruska invented th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intergenic Region
An intergenic region is a stretch of DNA sequences located between genes. Intergenic regions may contain functional elements and junk DNA. ''Inter''genic regions should not be confused with ''intra''genic regions (or introns), which are non-coding regions that are found ''within'' genes, especially within the genes of eukaryotic organisms. Properties and functions Intergenic regions may contain a number of functional DNA sequences such as promoters and regulatory elements, enhancers, spacers, and (in eukaryotes) centromeres. They may also contain origins of replication, scaffold attachment regions, and transposons and viruses. Non-functional DNA elements such as pseudogenes and repetitive DNA, both of which are types of junk DNA, can also be found in intergenic regions—although they may also be located within genes in introns. As all scientific knowledge is ultimately tentative—and in principle subject to revision given better evidence—it is possible s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |



.png)