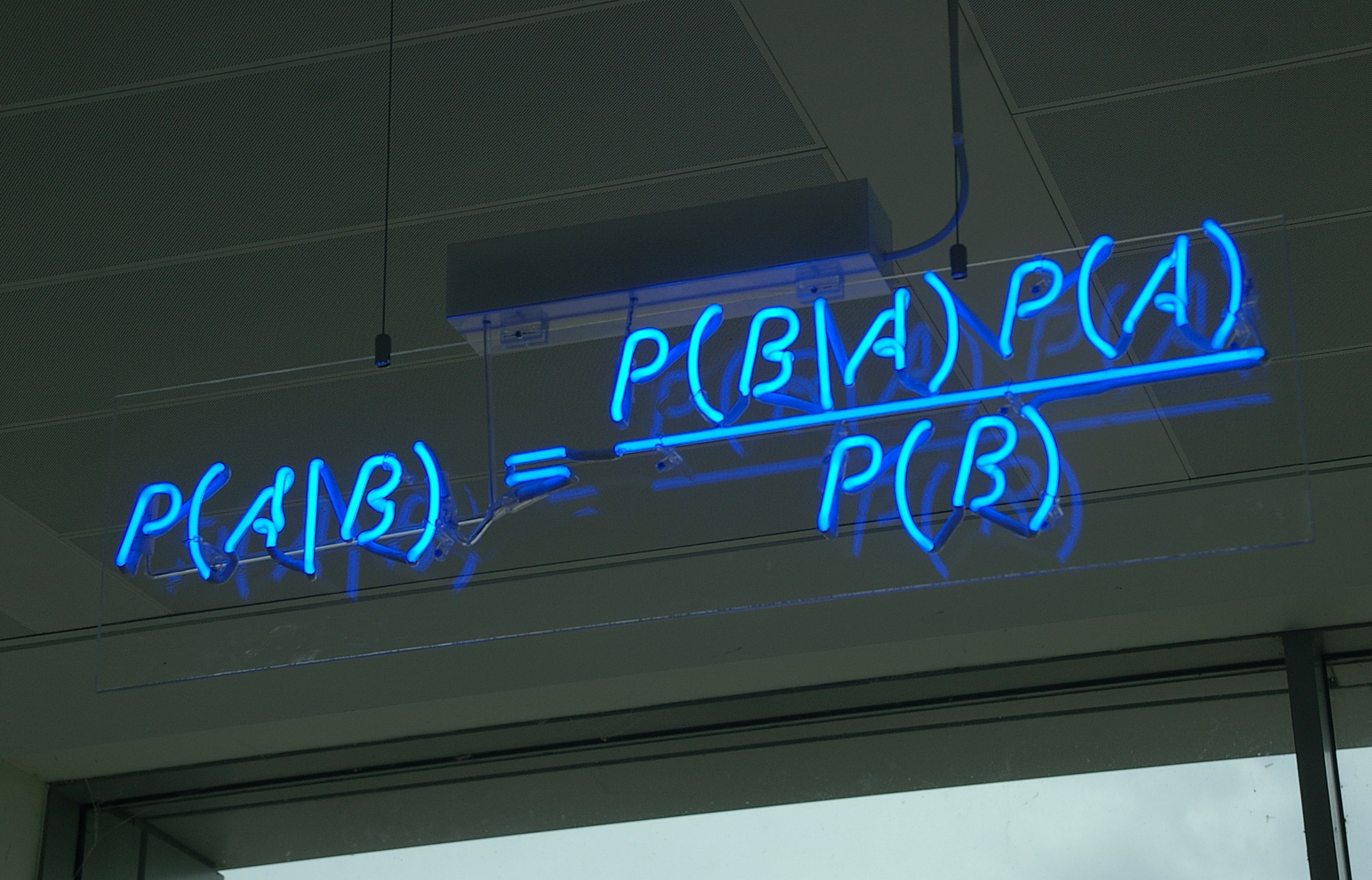|
Nexus File
The extensible NEXUS file format is widely used in bioinformatics. It stores information about taxa, morphological and molecular characters, distances, genetic codes, assumptions, sets, trees, etc. Several popular phylogenetic programs such as PAUP*,PAUP* — Phylogenetic Analysis Using Parsimony *and other methods ,MrBayes /ref> Mesquite,Mesquite: A modular system for ev ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for '' in silico'' analyses of biological queries using computational and statistical techniques. Bioinformatics includes biological studies that use computer programming as part of their methodology, as well as specific analysis "pipelines" that are repeatedly used, particularly in the field of genomics. Common uses of bioinformatics include the identification of candidates genes and single nucleotide polymorphisms (SNPs). Often, such identification is made with the aim to better understand the genetic basis of disease, unique adaptations, desirable properties (e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PAUP*
PAUP* (Phylogenetic Analysis Using Parsimony *and other methods) is a computational phylogenetics program for inferring evolutionary trees (phylogenies), written by David L. Swofford. Originally, as the name implies, PAUP only implemented parsimony, but from version 4.0 (when the program became known as PAUP*) it also supports distance matrix and likelihood methods. Version 3.0 ran on Macintosh computers and supported a rich, user-friendly graphical interface. Together with the program MacClade, with which it shares the NEXUS data format, PAUP* was the phylogenetic software of choice for many phylogenetists. Version 4.0 added support for Windows (graphical shell and command line) and Unix (command line only) platforms. However, the graphical user interface for the Macintosh does not support versions of Mac OS X higher than 10.14 (although a GUI for later versions of Mac OS is planned). PAUP* is also available as a plugin for Geneious. PAUP*, which now sports the self-referent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bayesian Inference In Phylogeny
Bayesian inference of phylogeny combines the information in the prior and in the data likelihood to create the so-called posterior probability of trees, which is the probability that the tree is correct given the data, the prior and the likelihood model. Bayesian inference was introduced into molecular phylogenetics in the 1990s by three independent groups: Bruce Rannala and Ziheng Yang in Berkeley, Bob Mau in Madison, and Shuying Li in University of Iowa, the last two being PhD students at the time. The approach has become very popular since the release of the MrBayes software in 2001, and is now one of the most popular methods in molecular phylogenetics. Bayesian inference of phylogeny background and bases Bayesian inference refers to a probabilistic method developed by Reverend Thomas Bayes based on Bayes' theorem. Published posthumously in 1763 it was the first expression of inverse probability and the basis of Bayesian inference. Independently, unaware of Bayes' work, Pierre- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SplitsTree
SplitsTree is a popular freeware program for inferring phylogenetic trees, phylogenetic networks, or, more generally, splits graphs, from various types of data such as a sequence alignment, a distance matrix or a set of trees. SplitsTree implements published methods such as split decomposition, neighbor-net, consensus networks, super networks methods or methods for computing hybridization or simple recombination networks. It uses the NEXUS file format. The splits graph is defined using a special data block (SPLITS block). See also *Phylogenetic tree viewers *Dendroscope Dendroscope is an interactive computer software program written in Java for viewing Phylogenetic trees. This program is designed to view trees of all sizes and is very useful for creating figures. Dendroscope can be used for a variety of analyse ... * MEGAN References External links SplitsTree homepage(New Website for informations about SplitsTree)for the latest version (4.15) and manual (June 201 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comment (programming)
In computer programming, a comment is a programmer-readable explanation or ''annotation'' in the source code of a computer program. They are added with the purpose of making the source code easier for humans to understand, and are generally ignored by compilers and interpreters.Source code can be divided into ''program code'' (which consists of machine-translatable instructions); and ''comments'' (which include human-readable notes and other kinds of annotations in support of the program code). The syntax of comments in various programming languages varies considerably. Comments are sometimes also processed in various ways to generate documentation external to the source code itself by documentation generators, or used for integration with source code management systems and other kinds of external programming tools. The flexibility provided by comments allows for a wide degree of variability, but formal conventions for their use are commonly part of programming style guides. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Newick Format
In mathematics, Newick tree format (or Newick notation or New Hampshire tree format) is a way of representing graph-theoretical trees with edge lengths using parentheses and commas. It was adopted by James Archie, William H. E. Day, Joseph Felsenstein, Wayne Maddison, Christopher Meacham, F. James Rohlf, and David Swofford, at two meetings in 1986, the second of which was at Newick's restaurant in Dover, New Hampshire, US. The adopted format is a generalization of the format developed by Meacham in 1984 for the first tree-drawing programs in Felsenstein's PHYLIP package. Examples The following tree: could be represented in Newick format in several ways ((,)); ''no nodes are named'' (A,B,(C,D)); ''leaf nodes are named'' (A,B,(C,D)E)F; ''all nodes are named'' (:0.1,:0.2,(:0.3,:0.4):0.5); ''all but root node have a distance to parent'' (:0.1,:0.2,(:0.3,:0.4):0.5):0.0; ''all hav ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NeXML Format
NeXML is an exchange standard for representing phyloinformatic data. It was inspired by the widely used Nexus file format but uses XML to produce a more robust format for rich phylogenetic data. Advantages include syntax validation, semantic annotation, and web services. The format is broadly supported and has libraries in many popular programming languages for bioinformatics Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combi .... External linksNeXML Github References {{Bioinformatics Biological sequence format ...[...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PhyloXML
PhyloXML is an XML language for the analysis, exchange, and storage of phylogenetic trees (or networks) and associated data. The structure of phyloXML is described by XML Schema Definition (XSD) language. A shortcoming of current formats for describing phylogenetic trees (such as Nexus and Newick/New Hampshire) is a lack of a standardized means to annotate tree nodes and branches with distinct data fields (which in the case of a basic species tree might be: species names, branch lengths, and possibly multiple support values). Data storage and exchange is even more cumbersome in studies in which trees are the result of a reconciliation of some kind: * gene-function studies (requires annotation of nodes with taxonomic information as well as gene names, and possibly gene-duplication data) * evolution of host-parasite interactions (requires annotation of tree nodes with taxonomic information for both host and parasite) * phylogeographic studies (requires annotation of tree nodes with ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for '' in silico'' analyses of biological queries using computational and statistical techniques. Bioinformatics includes biological studies that use computer programming as part of their methodology, as well as specific analysis "pipelines" that are repeatedly used, particularly in the field of genomics. Common uses of bioinformatics include the identification of candidates genes and single nucleotide polymorphisms (SNPs). Often, such identification is made with the aim to better understand the genetic basis of disease, unique adaptations, desirable properties (e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biological Sequence Format
Biology is the scientific study of life. It is a natural science with a broad scope but has several unifying themes that tie it together as a single, coherent field. For instance, all organisms are made up of cells that process hereditary information encoded in genes, which can be transmitted to future generations. Another major theme is evolution, which explains the unity and diversity of life. Energy processing is also important to life as it allows organisms to move, grow, and reproduce. Finally, all organisms are able to regulate their own internal environments. Biologists are able to study life at multiple levels of organization, from the molecular biology of a cell to the anatomy and physiology of plants and animals, and evolution of populations.Based on definition from: Hence, there are multiple subdisciplines within biology, each defined by the nature of their research questions and the tools that they use. Like other scientists, biologists use the scientific metho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |