|
NT5C
5', 3'-nucleotidase, cytosolic, also known as 5'(3')-deoxyribonucleotidase, cytosolic type (cdN) or deoxy-5'-nucleotidase 1 (dNT-1), is an enzyme that in humans is encoded by the ''NT5C'' gene on chromosome 17. This gene encodes a nucleotidase that catalyzes the dephosphorylation of the 5' deoxyribonucleotides (dNTP) and 2'(3')-dNTP and ribonucleotides, but not 5' ribonucleotides. Of the different forms of nucleotidases characterized, this enzyme is unique in its preference for 5'-dNTP. It may be one of the enzymes involved in regulating the size of dNTP pools in cells. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Nov 2011] Structure cdN is one of seven 5' nucleotidases identified in humans, all of which differ in tissue specificity, subcellular location, primary structure and substrate (biochemistry), substrate specificity. Of the seven, the mitochondrial counterpart of cdN, NT5M, mdN, is the most closely related to cdN. Their ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotidase
A nucleotidase is a hydrolytic enzyme that catalyzes the hydrolysis of a nucleotide into a nucleoside and a phosphate. : A nucleotide + H2O = a nucleoside + phosphate For example, it converts adenosine monophosphate to adenosine, and guanosine monophosphate to guanosine. Nucleotidases have an important function in digestion in that they break down consumed nucleic acids. They can be divided into two categories, based upon the end that is hydrolyzed: * : 5'-nucleotidase - NT5C, NT5C1A, NT5C1B, NT5C2, NT5C3 * : 3'-nucleotidase - NT3 5'-Nucleotidases cleave off the phosphate from the 5' end of the sugar moiety. They can be classified into various kinds depending on their substrate preferences and subcellular localization. Membrane-bound 5'-nucleotidases display specificity toward adenosine monophosphates and are involved predominantly in the salvage of preformed nucleotides and in signal transduction cascades involving purinergic receptors. Soluble 5'-nucleotidases are all known t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NT5M
5',3'-nucleotidase, mitochondrial, also known as 5'(3')-deoxyribonucleotidase, mitochondrial (mdN) or deoxy-5'-nucleotidase 2 (dNT-2), is an enzyme that in humans is encoded by the ''NT5M'' gene. This gene encodes a 5' nucleotidase that localizes to the mitochondrial matrix. This enzyme dephosphorylates the 5'- and 2'(3')-phosphates of uracil and thymine deoxyribonucleotides. The gene is located within the Smith–Magenis syndrome region on chromosome 17. Structure The cDNA of mdN encodes a 25.9-kDa polypeptide, and the crystal structure of this enzymes reveals a 196-long amino acid sequence in the mature protein. The first 32 amino acids, which contain the mitochondrial targeting sequence, are removed during the processing of the premature protein for its import into the mitochondrial matrix. The enzyme is likely a dimer protein formed by the interaction of alpha and beta loops between the cores of the two monomers. Each monomer is composed of a large and small domain connect ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
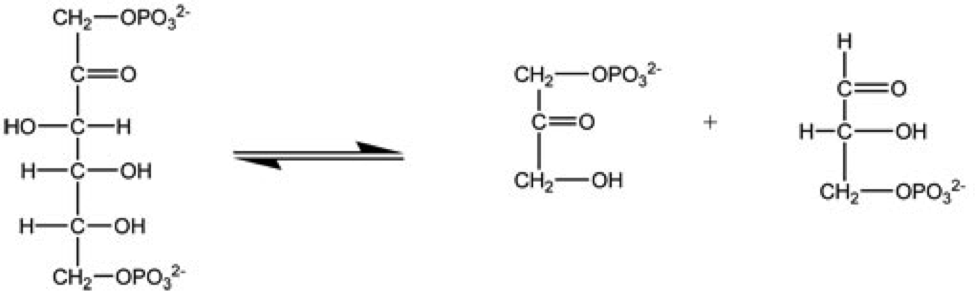
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
β-strand
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined versi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deoxyguanosine Monophosphate
Deoxyguanosine monophosphate (dGMP), also known as deoxyguanylic acid or deoxyguanylate in its conjugate acid and conjugate base forms, respectively, is a derivative of the common nucleic acid guanosine triphosphate (GTP), in which the –OH (hydroxyl) group on the 2' carbon on the nucleotide's pentose has been reduced to just a hydrogen atom (hence the "deoxy-" part of the name). It is used as a monomer in DNA. See also * Cofactor *Guanosine Guanosine (symbol G or Guo) is a purine nucleoside comprising guanine attached to a ribose ( ribofuranose) ring via a β-N9- glycosidic bond. Guanosine can be phosphorylated to become guanosine monophosphate (GMP), cyclic guanosine monophosphate ... * Nucleic acid References Nucleotides {{Biochemistry-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DTMP
Thymidine monophosphate (TMP), also known as thymidylic acid (conjugate base thymidylate), deoxythymidine monophosphate (dTMP), or deoxythymidylic acid (conjugate base deoxythymidylate), is a nucleotide that is used as a monomer in DNA. It is an ester of phosphoric acid with the nucleoside thymidine. dTMP consists of a phosphate group, the pentose sugar deoxyribose, and the nucleobase thymine. Unlike the other deoxyribonucleotides, thymidine monophosphate often does not contain the "deoxy" prefix in its name; nevertheless, its symbol often includes a "d" ("dTMP"). ''Dorland’s Illustrated Medical Dictionary'' provides an explanation of the nomenclature variation at its entry for thymidine. As a substituent, it is called by the prefix thymidylyl-. See also * DNA * Nucleoside * Nucleotide * Oligonucleotide * RNA Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DUMP
Dump generally refers to a place for disposal of solid waste, a rubbish dump, or landfill. The word has other uses alone or in combination, and may refer to: * Midden, historically a dump for domestic waste * Dump job, a term for criminal disposal of a corpse Arts, entertainment, and media * Dump (band), an alias for the solo recordings of James McNew * Dump, tape delay (broadcasting) * Dump months, times when, due to limited box office potential, new movies are generally perceived as being of low quality and limited appeal Computing and technology * Dump (Unix), a Unix program for backing up file systems * Storage dump, inaccurately but consistently referred to as a core dump in Unix-like systems, the recorded state of the working memory of a computer program at a specific time, generally when the program has terminated abnormally (crashed) * Database dump, a record of the table structure and/or the data from a database * ROM dump or ROM image, a record of the data in a ROM, E ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carbonyl
In organic chemistry, a carbonyl group is a functional group composed of a carbon atom double-bonded to an oxygen atom: C=O. It is common to several classes of organic compounds, as part of many larger functional groups. A compound containing a carbonyl group is often referred to as a carbonyl compound. The term carbonyl can also refer to carbon monoxide as a ligand in an inorganic or organometallic complex (a metal carbonyl, e.g. nickel carbonyl). The remainder of this article concerns itself with the organic chemistry definition of carbonyl, where carbon and oxygen share a double bond. Carbonyl compounds In organic chemistry, a carbonyl group characterizes the following types of compounds: Other organic carbonyls are urea and the carbamates, the derivatives of acyl chlorides chloroformates and phosgene, carbonate esters, thioesters, lactones, lactams, hydroxamates, and isocyanates. Examples of inorganic carbonyl compounds are carbon dioxide and carbonyl sulfide. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogen Bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a lone pair of electrons—the hydrogen bond acceptor (Ac). Such an interacting system is generally denoted , where the solid line denotes a polar covalent bond, and the dotted or dashed line indicates the hydrogen bond. The most frequent donor and acceptor atoms are the second-row elements nitrogen (N), oxygen (O), and fluorine (F). Hydrogen bonds can be intermolecular (occurring between separate molecules) or intramolecular (occurring among parts of the same molecule). The energy of a hydrogen bond depends on the geometry, the environment, and the nature of the specific donor and acceptor atoms and can vary between 1 and 40 kcal/mol. This makes them somewhat stronger than a van der Waals interaction, and weaker than fully covalent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Substrate Specificity
Chemical specificity is the ability of binding site of a macromolecule (such as a protein) to bind specific ligands. The fewer ligands a protein can bind, the greater its specificity. Specificity describes the strength of binding between a given protein and ligand. This relationship can be described by a dissociation constant, which characterizes the balance between bound and unbound states for the protein-ligand system. In the context of a single enzyme and a pair of binding molecules, the two ligands can be compared as stronger or weaker ligands (for the enzyme) on the basis of their dissociation constants. (A lower value corresponds to a stronger binding.) Specificity for a set of ligands is unrelated to the ability of an enzyme to catalyze a given reaction, with the ligand as a substrate. If a given enzyme has a high chemical specificity, this means that the set of ligands to which it binds is limited, such that neither binding events nor catalysis can occur at an appreciab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrophobic
In chemistry, hydrophobicity is the physical property of a molecule that is seemingly repelled from a mass of water (known as a hydrophobe). In contrast, hydrophiles are attracted to water. Hydrophobic molecules tend to be nonpolar and, thus, prefer other neutral molecules and nonpolar solvents. Because water molecules are polar, hydrophobes do not dissolve well among them. Hydrophobic molecules in water often cluster together, forming micelles. Water on hydrophobic surfaces will exhibit a high contact angle. Examples of hydrophobic molecules include the alkanes, oils, fats, and greasy substances in general. Hydrophobic materials are used for oil removal from water, the management of oil spills, and chemical separation processes to remove non-polar substances from polar compounds. Hydrophobic is often used interchangeably with lipophilic, "fat-loving". However, the two terms are not synonymous. While hydrophobic substances are usually lipophilic, there are exceptions, suc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aromatic
In chemistry, aromaticity is a chemical property of cyclic ( ring-shaped), ''typically'' planar (flat) molecular structures with pi bonds in resonance (those containing delocalized electrons) that gives increased stability compared to saturated compounds having single bonds, and other geometric or connective non-cyclic arrangements with the same set of atoms. Aromatic rings are very stable and do not break apart easily. Organic compounds that are not aromatic are classified as aliphatic compounds—they might be cyclic, but only aromatic rings have enhanced stability. The term ''aromaticity'' with this meaning is historically related to the concept of having an aroma, but is a distinct property from that meaning. Since the most common aromatic compounds are derivatives of benzene (an aromatic hydrocarbon common in petroleum and its distillates), the word ''aromatic'' occasionally refers informally to benzene derivatives, and so it was first defined. Nevertheless, many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |




.png)