|
NFATC2
Nuclear factor of activated T-cells, cytoplasmic 2 is a protein that in humans is encoded by the ''NFATC2'' gene. Function This gene is a member of the nuclear factor of activated T cells (NFAT) family. The product of this gene is a DNA-binding protein with a REL-homology region (RHR) and an NFAT-homology region (NHR). This protein is present in the cytosol and only translocates to the nucleus upon T cell receptor (TCR) stimulation, where it becomes a member of the nuclear factors of activated T cells transcription complex. This complex plays a central role in inducing gene transcription during the immune response. Alternate transcriptional splice variants, encoding different isoforms, have been characterized. Clinical significance Translocation forming an in frame fusions product between EWSR1 gene and the NFATc2 gene has been described in bone tumor with a Ewing sarcoma-like clinical appearance. The translocation breakpoint led to the loss of the controlling elements of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NFAT
Nuclear factor of activated T-cells (NFAT) is a family of transcription factors shown to be important in immune response. One or more members of the NFAT family is expressed in most cells of the immune system. NFAT is also involved in the development of cardiac, skeletal muscle, and nervous systems. NFAT was first discovered as an activator for the transcription of IL-2 in T cells (as a regulator of T cell immune response) but has since been found to play an important role in regulating many more body systems. NFAT transcription factors are involved in many normal body processes as well as in development of several diseases, such as inflammatory bowel diseases and several types of cancer. NFAT is also being investigated as a drug target for several different disorders. Family members The NFAT transcription factor family consists of five members NFATc1, NFATc2, NFATc3, NFATc4, and NFAT5. NFATc1 through NFATc4 are regulated by calcium signalling, and are known as the classical mem ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MEF2D
Myocyte-specific enhancer factor 2D is a protein that in humans is encoded by the ''MEF2D'' gene. Interactions MEF2D has been shown to interact with: * CABIN1, * EP300, * MAPK7, * Myocyte-specific enhancer factor 2A, * NFATC2 * Sp1 transcription factor, and * YWHAQ. See also * Mef2 In the field of molecular biology, myocyte enhancer factor-2 (Mef2) proteins are a family of transcription factors which through control of gene expression are important regulators of cellular differentiation and consequently play a critical rol ... References Further reading * * * * * * * * * * * * * * * * External links * {{Transcription factors, g4 Transcription factors ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EP300
Histone acetyltransferase p300 also known as p300 HAT or E1A-associated protein p300 (where E1A = adenovirus early region 1A) also known as EP300 or p300 is an enzyme that, in humans, is encoded by the ''EP300'' gene. It functions as histone acetyltransferase that regulates transcription of genes via chromatin remodeling by allowing histone proteins to wrap DNA less tightly. This enzyme plays an essential role in regulating cell growth and division, prompting cells to mature and assume specialized functions (differentiate), and preventing the growth of cancerous tumors. The p300 protein appears to be critical for normal development before and after birth. The EP300 gene is located on the long (q) arm of the human chromosome 22 at position 13.2. This gene encodes the adenovirus E1A-associated cellular p300 transcriptional co-activator protein. EP300 is closely related to another gene, CREB binding protein, which is found on human chromosome 16. Function p300 HAT functions ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
IRF4
Interferon regulatory factor 4 (IRF4) also known as ''MUM1'' is a protein that in humans is encoded by the ''IRF4'' gene, located at 6p25-p23. IRF4 functions as a key regulatory transcription factor in the development of human immune cells.Nam S, Lim J-S (2016). "Essential role of interferon regulatory factor 4 (IRF4) in immune cell development." ''Arch. Pharm. Res''. 39: 1548–1555doi:10.1007/s12272-016-0854-1Shaffer AL, Tolga Emre NC, Romesser PB, Staudt LM (2009). "IRF4: Immunity. Malignancy! Therapy?" ''Clinical Cancer Research''. 15 (9): 2954-2961doi:10.1158/1078-0432.CCR-08-1845/ref> The expression of IRF4 is essential for the differentiation of T lymphocytes and B lymphocytes as well as certain myeloid cells. The ''MUM1'' symbol is polysemous; although it is an older synonym for ''IRF4'' (HGNC:6119), it is also the current HGNC official symbol for melanoma associated antigen (mutated) 1 (HGNC:29641; located at 19p13.3). Dysregulation of the ''IRF4'' gene can result in ''IRF ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Kinase Mζ
Protein kinase C, zeta (PKCζ), also known as PRKCZ, is a protein in humans that is encoded by the ''PRKCZ'' gene. The PRKCZ gene encodes at least two alternative transcripts, the full-length PKCζ and an N-terminal truncated form PKMζ. PKMζ is thought to be responsible for maintaining long-term memories in the brain. The importance of PKCζ in the creation and maintenance of long-term potentiation was first described by Todd Sacktor and his colleagues at the SUNY Downstate Medical Center in 1993. Structure PKC-zeta has an N-terminal regulatory domain, followed by a hinge region and a C-terminal catalytic domain. Second messengers stimulate PKCs by binding to the regulatory domain, translocating the enzyme from cytosol to membrane, and producing a conformational change that removes auto-inhibition of the PKC catalytic protein kinase activity. PKM-zeta, a brain-specific isoform of PKC-zeta generated from an alternative transcript, lacks the regulatory region of full-length PKC-z ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA po ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA-binding Protein
DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for DNA#Base pairing, single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair. However, there are some known minor groove DNA-binding ligands such as netropsin, distamycin, Hoechst 33258, pentamidine, DAPI and others. Examples DNA-binding proteins include transcription factors which Gene modulation, modulate the process of transcription, various polymerases, nucleases which cleave DNA molecules, and histones which are involved in chromosome packaging and transcription in the cell nucleus. DNA-binding proteins can incorporate such domains as the zinc finger, the helix-turn-helix, and the leucine zipper (among many others) that facilitate binding to nucleic acid. There are also more unusual examples such as TAL effector, transcription activa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
REL-homology Region
The Rel homology domain (RHD) is a protein domain found in a family of eukaryotic transcription factors, including both NF-κB and NFAT, among others. Some of these transcription factors appear to form multi-protein DNA-bound complexes. Phosphorylation of the RHD appears to play a role in the regulation of some of these transcription factors, acting to modulate the expression of their target genes. The RHD is composed of two immunoglobulin-like beta barrel subdomains that grip the DNA in the major groove. The N-terminal specificity domain resembles the core domain of the p53 transcription factor, and contains a recognition loop that interacts with DNA bases. In the case of NF-κB, the C-terminal dimerization subdomain determines dimerization propensity with other proteins in the NF-κB Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) is a protein complex that controls transcription of DNA, cytokine production and cell survival. NF-κB is found in a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
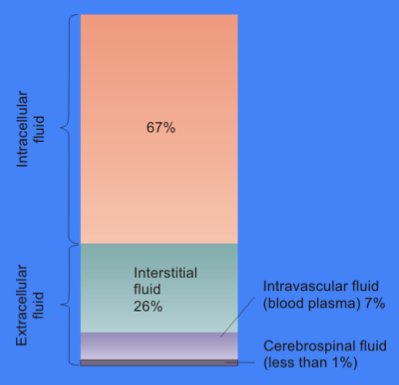
Cytosol
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells (intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into many compartments. In the eukaryotic cell, the cytosol is surrounded by the cell membrane and is part of the cytoplasm, which also comprises the mitochondria, plastids, and other organelles (but not their internal fluids and structures); the cell nucleus is separate. The cytosol is thus a liquid matrix around the organelles. In prokaryotes, most of the chemical reactions of metabolism take place in the cytosol, while a few take place in membranes or in the periplasmic space. In eukaryotes, while many metabolic pathways still occur in the cytosol, others take place within organelles. The cytosol is a complex mixture of substances dissolved in water. Although water forms the large majority of the cytosol, its structure and prope ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ewing Sarcoma Breakpoint Region 1
RNA-binding protein EWS is a protein that in humans is encoded by the ''EWSR1'' gene on human chromosome 22, specifically 22q12.2. It is one of 3 proteins in the FET protein family. The q22.2 region of chromosome 22 encodes the N-terminal transactivation domain of the EWS protein and that region may become joined to one of several other chromosomes which encode various transcription factors, see and the FET protein family. The expression of a chimeric protein with the EWS transactivation domain fused to the DNA binding region of a transcription factor generates a powerful oncogenic protein causing Ewing sarcoma and other members of the Ewing family of tumors. These translocations can occur due to chromoplexy, a burst of complex chromosomal rearrangements seen in cancer cells. The normal EWS gene encodes an RNA binding protein closely related to FUS (gene) and TAF15, all of which have been associated to amyotrophic lateral sclerosis. Interactions The EWS protein has been shown to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |