|
Nuclear Receptor Coregulators
Nuclear receptor coregulatorsThis article is based on the transcript of an animated course on the Nuclear Receptor Signaling Atlas (NURSA) website entitled "Nuclear receptor signaling: concepts and models". Permission to re-use this material was obtained from the original copyright holder, the Nuclear Receptor Signaling Atlas, on August 25th 2008. are a class of transcription coregulators that have been shown to be involved in any aspect of signaling by any member of the nuclear receptor superfamily. A comprehensive database of coregulators for nuclear receptors and other transcription factors was previously maintained at the Nuclear Receptor Signaling Atlas website which has since been replaced by thSignaling Pathways Projectwebsite. Introduction The ability of nuclear receptors to alternate between activation and repression in response to specific molecular cues, is now known to be attributable in large part to a diverse group of cellular factors, collectively termed coregulator ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Coregulators
In molecular biology and genetics, transcription coregulators are proteins that interact with transcription factors to either activate or repress the transcription of specific genes. Transcription coregulators that activate gene transcription are referred to as coactivators while those that repress are known as corepressors. The mechanism of action of transcription coregulators is to modify chromatin structure and thereby make the associated DNA more or less accessible to transcription. In humans several dozen to several hundred coregulators are known, depending on the level of confidence with which the characterisation of a protein as a coregulator can be made. One class of transcription coregulators modifies chromatin structure through covalent modification of histones. A second ATP dependent class modifies the conformation of chromatin. Histone acetyltransferases Nuclear DNA is normally tightly wrapped around histones rendering the DNA inaccessible to the general transcr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
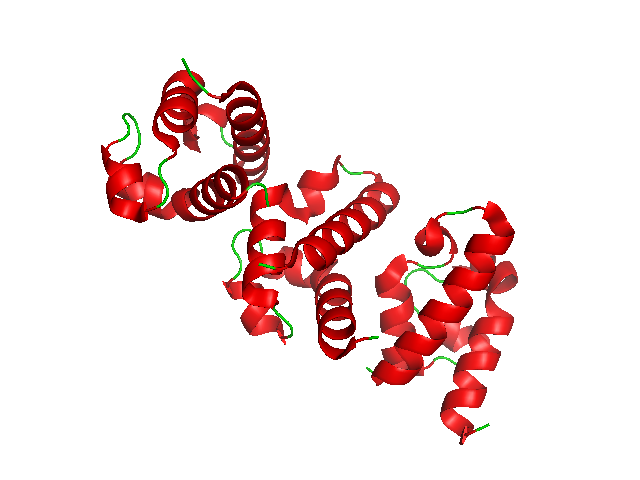
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astbu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mediator (coactivator)
Mediator is a multiprotein complex that functions as a transcriptional coactivator in all eukaryotes. It was discovered in 1990 in the lab of Roger D. Kornberg, recipient of the 2006 Nobel Prize in Chemistry. Mediator complexes interact with transcription factors and RNA polymerase II. The main function of mediator complexes is to transmit signals from the transcription factors to the polymerase. Mediator complexes are variable at the evolutionary, compositional and conformational levels. The first image shows only one "snapshot" of what a particular mediator complex might be composed of, but it certainly does not accurately depict the conformation of the complex ''in vivo''. During evolution, mediator has become more complex. The yeast ''Saccharomyces cerevisiae'' (a simple eukaryote) is thought to have up to 21 subunits in the core mediator (exclusive of the CDK module), while mammals have up to 26. Individual subunits can be absent or replaced by other subunits under differ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DDX17
Probable ATP-dependent RNA helicase DDX17 (p72) is an enzyme that in humans is encoded by the ''DDX17'' gene. Function DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. This gene encodes a DEAD box protein, which is an ATPase activated by a variety of RNA species but not by dsDNA. This protein and that encoded by DDX5 gene are more closely related to each other than to any other member of the DEAD box family. Alternative splicing of this gene generates 2 transcript variants encoding different isoforms with the longer transcript reported to also initiate tran ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Helicase
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands (hence '' helic- + -ase''), using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases. The human genome codes for 95 non-redundant helicases: 64 RNA helicases and 31 DNA helicases. Many cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair, and ribosome biogenesis involve the separation of nucleic acid strands that necessitates the use of helicases. Some specialized helicases are also involved in sensing of viral nucleic acids during infection and fulfill a immunological function. Function Helicases are oft ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CDC25B
M-phase inducer phosphatase 2 is an enzyme that in humans is encoded by the ''CDC25B'' gene. CDC25B is a member of the CDC25 family of phosphatases. CDC25B activates the cyclin dependent kinase CDC2 by removing two phosphate groups and it is required for entry into mitosis. CDC25B shuttles between the nucleus and the cytoplasm due to nuclear localization and nuclear export signals. The protein is nuclear in the M and G1 phases of the cell cycle and moves to the cytoplasm during S and G2. CDC25B has oncogenic properties, although its role in tumor formation has not been determined. Multiple transcript variants for this gene exist. Interactions CDC25B has been shown to interact with MAPK14, Casein kinase 2, alpha 1, CHEK1, MELK, Estrogen receptor alpha, YWHAB, YWHAZ, YWHAH and YWHAE 14-3-3 protein epsilon is a protein that in humans is encoded by the ''YWHAE'' gene. Function This gene product belongs to the 14-3-3 family of proteins which mediate signal transduction by bindin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SRA1
Steroid receptor RNA activator 1 also known as steroid receptor RNA activator protein (SRAP) is a protein that in humans is encoded by the ''SRA1'' gene. The mRNA transcribed from the SRA1 gene is a component of the ribonucleoprotein complex containing NCOA1. This functional RNA also encodes a protein. Function This gene is involved in transcriptional coactivation by steroid receptor. There is currently data suggesting this gene encodes both a non-coding RNA that functions as part of a ribonucleoprotein complex and a protein coding mRNA. Increased expression of both the transcript and the protein is associated with cancer. Interactions SRA1 has been shown to interact with: * Estrogen receptor alpha, * DDX17, * Nuclear receptor coactivator 2, and * SPEN Msx2-interacting protein is a protein that in humans is encoded by the ''SPEN'' gene. This gene encodes a hormone inducible transcriptional repressor. Repression of transcription by this gene product can occur through ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PRMT1
Protein arginine N-methyltransferase 1 is an enzyme that in humans is encoded by the ''PRMT1'' gene. The HRMT1L2 gene encodes a protein arginine methyltransferase that functions as a histone methyltransferase specific for histone H4. Function PRMT1 gene encodes for the protein arginine methyltransferase that functions as a histone methyltransferase specific for histone H4 in eukaryotic cells. Specifically altering histone H4 in eukaryotes gives it the ability to remodel chromatin acting as a post-translational modifier. Through regulation of gene expression, arginine methyltransferases control the cell cycle and death of eukaryotic cells. Reaction pathway While all PRMT enzymes catalyze the methylation of arginine residues in proteins, PRMT1 is unique in that is catalyzes the formation of asymmetric dimethylarginine as opposed to the PRMT2 that catalyzes the formation of symmetrically dimethylated arginine. Individual PRMT utilize ''S''-adenosyl-L-methionine (SAM) as th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methylase
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding ''S''-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the lea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SMARCA4
Transcription activator BRG1 also known as ATP-dependent chromatin remodeler SMARCA4 is a protein that in humans is encoded by the ''SMARCA4'' gene. Function The protein encoded by this gene is a member of the SWI/SNF family of proteins and is similar to the brahma protein of Drosophila. Members of this family have helicase and ATPase activities and are thought to regulate transcription of certain genes by altering the chromatin structure around those genes. The encoded protein is part of the large ATP-dependent chromatin remodeling complex SWI/SNF, which is required for transcriptional activation of genes normally repressed by chromatin. In addition, this protein can bind BRCA1, as well as regulate the expression of the tumorigenic protein CD44. BRG1 works to activate or repress transcription. Having functional BRG1 is important for development past the pre-implantation stage. Without having a functional BRG1, exhibited with knockout research, the embryo will not hatch out ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SWI/SNF
In molecular biology, SWI/SNF (SWItch/Sucrose Non-Fermentable), is a subfamily of ATP-dependent chromatin remodeling complexes, which is found in eukaryotes. In other words, it is a group of proteins that associate to remodel the way DNA is packaged. This complex is composed of several proteins – products of the SWI and SNF genes (, /, , , ), as well as other polypeptides. It possesses a DNA-stimulated ATPase activity that can destabilize histone-DNA interactions in reconstituted nucleosomes in an ATP-dependent manner, though the exact nature of this structural change is unknown. The SWI/SNF subfamily provides crucial nucleosome rearrangement, which is seen as ejection and/or sliding. The movement of nucleosomes provides easier access to the chromatin, allowing genes to be activated or repressed. The human analogs of SWI/SNF are " BRG1- or BRM-associated factors", or BAF (SWI/SNF-A) and "Polybromo-associated BAF", which is also known as PBAF (SWI/SNF-B). There are also ''Dros ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UBE3A
Ubiquitin-protein ligase E3A (UBE3A) also known as E6AP ubiquitin-protein ligase (E6AP) is an enzyme that in humans is encoded by the ''UBE3A'' gene. This enzyme is involved in targeting proteins for degradation within cells. Protein degradation is a normal process that removes damaged or unnecessary proteins and helps maintain the normal functions of cells. Ubiquitin protein ligase E3A attaches a small marker protein called ubiquitin to proteins that should be degraded. Cellular structures called proteasomes recognize and digest proteins tagged with ubiquitin. Both copies of the UBE3A gene are active in most of the body's tissues. In most neurons, however, only the copy inherited from a person's mother (the maternal copy) is normally active; this is known as paternal imprinting. Recent evidence shows that at least some glial cells and neurons may exhibit biallelic expression of UBE3A. Further work is thus needed to delineate a complete map of UBE3A imprinting in humans and mod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |